Python预测分析(7):用Python构建集成模型
Python工具包的易用性、所能达到的准确性、训练所需的时间等等
7.1 用Python集成方法工具包解决回归问题
构建随机森林模型来预测红酒口感:scikit-learn中RandomForestRegressor的类构造函数如下
sklearn.ensemble.RandomForestRegressor(n_estimators=10, criterion='mse', max_depth=None, min_samples_split=2, min_samples_leaf=1, max_features='auto', max_leaf_nodes=None, bootstrap=True, oob_score=False, n_jobs=1, random_state=None, verbose=0, min_density=None, compute_importances=None)
n_estimators: 决策树的数目。比较好的尝试是100~500
max_depth: 如果设置为None,决策树就会持续增长。
min_samples_split:当节点含有的数据实例少于min_samples_split时,此节点不再分割。
min_samples_leaf:如果分割导致节点拥有的数据实例少于min_samples_leaf,分割就不会进行。
max_features:如果是整型,则为属性个数,如果是浮点型,则为百分比。
random_state:随机数生成器
类的几个属性
feature_importances: 属性重要程度
fit(XTrain, yTrain, sample_weight=None)
predict(XTest)
__author__ = 'mike-bowles'
import urllib.request as urllib2
import numpy
from sklearn.cross_validation import train_test_split
from sklearn import ensemble
from sklearn.metrics import mean_squared_error
import random
from math import sqrt
import pylab as plot
import csv
from io import StringIO as pyIO
target_url = ("http://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-red.csv")
data = urllib2.urlopen(target_url)
data = data.read().decode('utf-8')
dataFile = pyIO(data)
csvReader = csv.reader(dataFile)
xList = []
labels = []
names = []
firstLine = True
for line in csvReader:
if firstLine:
print(line)
firstLine = False
continue
row = line[0].strip().split(";")
labels.append(float(row[-1]))
row.pop()
floatRow = [float(num) for num in row]
xList.append(floatRow)
nrows = len(xList)
ncols = len(xList[0])
X = numpy.array(xList)
y = numpy.array(labels)
wineNames = numpy.array(names)
xTrain, xTest, yTrain, yTest = train_test_split(X, y, test_size=0.30, random_state=531)
# see how the mse changes
mseOos = []
nTreeList = range(50, 500, 10)
for iTrees in nTreeList:
depth = None
maxFeat = 4 # try tweaking
wineRFModel = ensemble.RandomForestRegressor(n_estimators=iTrees, max_depth=depth, max_features=maxFeat, oob_score=False, random_state=531)
wineRFModel.fit(xTrain, yTrain)
prediction = wineRFModel.predict(xTest)
mseOos.append(mean_squared_error(yTest, prediction))
print("MSE")
print(mseOos[-1])
plot.plot(nTreeList, mseOos)
plot.ylabel('Number of Trees in Ensemble')
plot.ylim([0.0, 1.1*max(mseOos)])
plot.show()
#
featureImportance = wineRFModel.feature_importances_
# scale by max importance
featureImportance = featureImportance/featureImportance.max()
sorted_idx = numpy.argsort(featureImportance)
barPos = numpy.arange(sorted_idx.shape[0]) + .5
plot.barh(barPos, featureImportance[sorted_idx], align="center")
plot.yticks(barPos, wineNames[sorted_idx])
plot.xlabel('Variable Importance')
plot.show()用梯度提升预测红酒品质:sklearn中GradientBoostingRegressor的类构造函数
sklearn.ensemble.GradientBoostingRegressor(loss='ls', learning_rate=0.1, n_estimators=100, subsample=1.0, min_samples_split=2, min_samples_leaf=1, max_depth=3, init=None, random_state=None, max_features=None, alpha=0.9, verbose=0, max_leaf_nodes=None, warm_start=False)
Loss:损失函数,ls(最小均方误差)lad(最小平均绝对误差)huber(胡贝尔误差)quantile(分位数回归)Learning_rate
N_estimators:决策树数目
Subsample:算法发明人建议取0.5
Max_depth:
Max_features:属性数量
Warm_start:为True,fit函数将从上次训练停止的地方开始
属性
Feature_importance:
Train_score:训练阶段对决策树依次训练时的误差
方法
Fit(XTrain, yTrain, monitor=None)
Predict(X)
Staged_predict(x):
__author__ = 'mike-bowles'
import urllib.request as urllib2
import numpy
from sklearn.cross_validation import train_test_split
from sklearn import ensemble
from sklearn.metrics import mean_squared_error
import random
from math import sqrt
import pylab as plot
import csv
from io import StringIO as pyIO
target_url = ("http://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-red.csv")
data = urllib2.urlopen(target_url)
data = data.read().decode('utf-8')
dataFile = pyIO(data)
csvReader = csv.reader(dataFile)
xList = []
labels = []
names = []
firstLine = True
for line in csvReader:
if firstLine:
print(line)
firstLine = False
continue
row = line[0].strip().split(";")
labels.append(float(row[-1]))
row.pop()
floatRow = [float(num) for num in row]
xList.append(floatRow)
nrows = len(xList)
ncols = len(xList[0])
X = numpy.array(xList)
y = numpy.array(labels)
wineNames = numpy.array(names)
xTrain, xTest, yTrain, yTest = train_test_split(X, y, test_size=0.30, random_state=531)
nEst = 2000
depth = 7
learnRate = 0.01
subSamp = 0.5
wineGBMModel = ensemble.GradientBoostingRegressor(n_estimators=nEst, max_depth=depth, learning_rate=learnRate, subsample=subSamp, loss='ls')
wineGBMModel.fit(xTrain, yTrain)
# compute mse on test set
msError = []
predictions = wineGBMModel.staged_predict(xTest)
for p in predictions:
msError.append(mean_squared_error(yTest, p))
print("MSE")
print(min(msError))
print(msError.index(min(msError)))
plot.figure()
plot.plot(range(1, nEst+1), wineGBMModel.train_score_, label="Training Set MSE")
plot.plot(range(1, nEst+1), msError, label="Test Set MSE")
plot.legend(loc="upper right")
plot.xlabel("Number of Trees in Ensemble")
plot.ylabel("Mean Squared Error")
plot.show()
featureImportance = wineGBMModel.feature_importances_
# scale by max importance
featureImportance = featureImportance/featureImportance.max()
sorted_idx = numpy.argsort(featureImportance)
barPos = numpy.arange(sorted_idx.shape[0]) + .5
plot.barh(barPos, featureImportance[sorted_idx], align="center")
plot.yticks(barPos, wineNames[sorted_idx])
plot.xlabel('Variable Importance')
plot.subplots_adjust(left=0.2, right=0.9, top=0.9, bottom=0.1)
plot.show()用Bagging来预测红酒口感:见第6章
7.3 Python集成方法引入非数值属性
非数值属性是指那些某几个离散非数值型的属性。人口普查记录就含有大量的非数值属性,已婚、单身或离异。
对鲍鱼性别属性编码引入Python随机森林回归方法:
__author__ = 'mike-bowles'
import urllib.request as urllib2
import numpy
import matplotlib.pyplot as plot
from sklearn.cross_validation import train_test_split
from sklearn import ensemble
from sklearn.metrics import mean_squared_error
import csv
from io import StringIO as pyIO
target_url = ("http://archive.ics.uci.edu/ml/machine-learning-databases/abalone/abalone.data")
data = urllib2.urlopen(target_url)
data = data.read().decode('utf-8')
dataFile = pyIO(data)
csvReader = csv.reader(dataFile)
xList = []
labels = []
names = []
for line in csvReader:
#print(line)
row = line
labels.append(float(row.pop()))
xList.append(row)
xCoded = []
for row in xList:
codedSex = [0.0, 0.0]
if row[0]=='M': codedSex[0] = 1.0
if row[0]=='F': codedSex[1] = 1.0
numRow = [float(row[i]) for i in range(1, len(row))]
rowCoded = list(codedSex) + numRow
xCoded.append(rowCoded)
abaloneNames = numpy.array(['Sex1', 'Sex2', 'Length', 'Diameter', 'Height', 'Whole weight', 'Shucked weight', 'Viscera weight', 'Shell weight', 'Rings'])
nrows = len(xCoded)
ncols = len(xCoded[1])
X = numpy.array(xCoded)
y = numpy.array(labels)
xTrain, xTest, yTrain, yTest = train_test_split(X, y, test_size=0.30, random_state=531)
mseOos = []
nTreeList = range(50, 500, 10)
for iTrees in nTreeList:
depth = None
maxFeat = 4
abaloneRFModel = ensemble.RandomForestRegressor(n_estimators=iTrees, max_depth=depth, max_features=maxFeat, oob_score=False, random_state=531)
abaloneRFModel.fit(xTrain, yTrain)
# Accumulate mse
prediction = abaloneRFModel.predict(xTest)
mseOos.append(mean_squared_error(yTest, prediction))
print("MSE")
print(mseOos[-1])
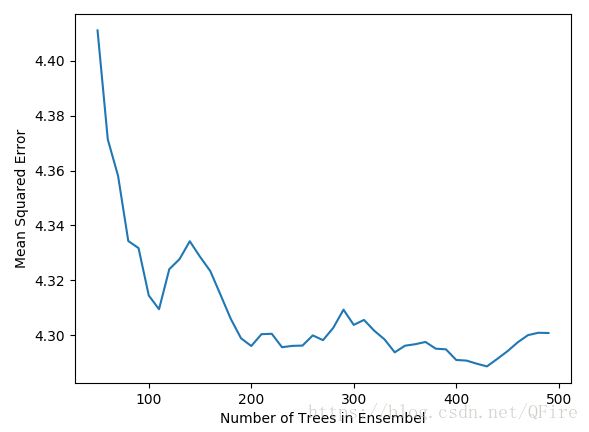
plot.plot(nTreeList, mseOos)
plot.xlabel('Number of Trees in Ensembel')
plot.ylabel('Mean Squared Error')
plot.show()
featureImportance = abaloneRFModel.feature_importances_
featureImportance = featureImportance/featureImportance.max()
sortedIdx = numpy.argsort(featureImportance)
barPos = numpy.arange(sortedIdx.shape[0]) + 0.5
plot.barh(barPos, featureImportance[sortedIdx], align="center")
plot.yticks(barPos, abaloneNames[sortedIdx])
plot.xlabel('Variable Importance')
plot.subplots_adjust(left=0.2, right=0.9, top=0.9, bottom=0.1)
plot.show()7.4 用Python集成方法解决二分类问题
用Python随机森林方法探测未爆炸的水雷:RandomForestClassifier类
sklearn.ensemble.RandomForestClassifier(n_estimators=10, criterion='gini', max_depth=None, min_samples_split=2, min_samples_leaf=1, max_features='auto', max_leaf_nodes=None, bootstrap=True, oob_score=False, n_jobs=1, random_state=None, verbose=0, min_density=None, compute_importances=None)
Criterion:可能的取值:Gini(基尼不纯度)Entropy(基于熵的信息增益)
为了生成曲线下面积(area under the curve, AUC),可能想获取接受者操作特征曲线(receiver operating curve, ROC)及其概率以保证精确度。如果想计算误分类率,则需要将概率转换为类别的预测。
Fit(X, y, sample_weight=None) 标签是从0到nClass-1的整数
Predict(X)生成一个单列的数组
Predict_proba(X)产生一个二维数组,对应类别的概率
Predict_log_proba(X)产生一个二维数组,对应的概率的log值
__author__ = 'mike-bowles'
import urllib.request as urllib2
import numpy
import matplotlib.pyplot as plot
from sklearn.cross_validation import train_test_split
from sklearn import ensemble
from sklearn.metrics import mean_squared_error, roc_auc_score, roc_curve
import csv
from io import StringIO as pyIO
from math import sqrt, fabs, exp
target_url = ("http://archive.ics.uci.edu/ml/machine-learning-databases/undocumented/connectionist-bench/sonar/sonar.all-data")
data = urllib2.urlopen(target_url)
data = data.read().decode('utf-8')
dataFile = pyIO(data)
csvReader = csv.reader(dataFile)
xList = []
labels = []
names = []
for line in csvReader:
#print(line)
row = line
xList.append(row)
xCoded = []
xNum = []
for row in xList:
lastCol = row.pop()
if lastCol == 'M':
labels.append(1)
else:
labels.append(0)
attrRow = [float(elt) for elt in row]
xNum.append(attrRow)
nrows = len(xNum)
ncols = len(xNum[1])
X = numpy.array(xNum)
y = numpy.array(labels)
rocksVMinesNames = numpy.array(['V'+str(i) for i in range(ncols)])
xTrain, xTest, yTrain, yTest = train_test_split(X, y, test_size=0.30, random_state=531)
auc = []
nTreeList = range(50, 2000, 50)
for iTrees in nTreeList:
depth = None
maxFeat = 8
rocksVMinesRMModel = ensemble.RandomForestClassifier(n_estimators=iTrees, max_depth=depth, max_features=maxFeat, oob_score=False, random_state=531)
rocksVMinesRMModel.fit(xTrain, yTrain)
prediction = rocksVMinesRMModel.predict_proba(xTest)
aucCalc = roc_auc_score(yTest, prediction[:, 1:2])
auc.append(aucCalc)
print("AUC")
print(auc[-1])
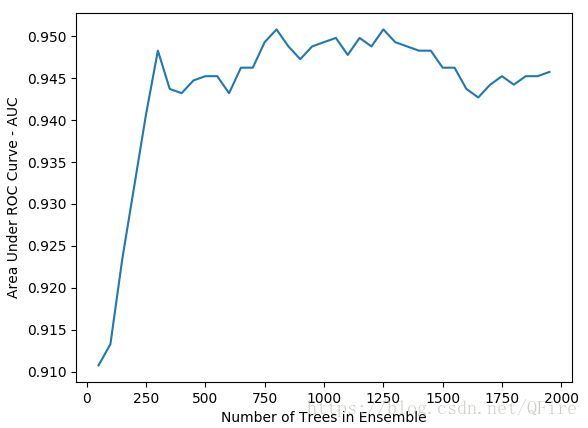
plot.plot(nTreeList, auc)
plot.xlabel('Number of Trees in Ensemble')
plot.ylabel('Area Under ROC Curve - AUC')
plot.show()
featureImportance = rocksVMinesRMModel.feature_importances_
featureImportance = featureImportance/featureImportance.max()
sortedIdx = numpy.argsort(featureImportance)
barPos = numpy.arange(sortedIdx.shape[0]) + 0.5
plot.barh(barPos, featureImportance[sortedIdx], align="center")
plot.yticks(barPos, rocksVMinesNames[sortedIdx])
plot.xlabel('Variable Importance')
#plot.subplots_adjust(left=0.2, right=0.9, top=0.9, bottom=0.1)
plot.show()
# plot best version of ROC curve
fpr, tpr, thresh = roc_curve(yTest, list(prediction[:, 1:2]))
ctClass = [i*0.01 for i in range(101)]
plot.plot(ctClass, ctClass, linestyle=":")
plot.xlabel('False Positive Rate')
plot.ylabel('True Positive Rate')
plot.show()
idx25 = int(len(thresh) * 0.25)
idx50 = int(len(thresh) * 0.50)
idx75 = int(len(thresh) * 0.75)
totalPts = len(yTest)
P = sum(yTest)
N = totalPts - P
print("")
print("Confusion Matrices for Different Threshold Values")
# 25th
idx = idx25
TP = tpr[idx] * P; FN = P - TP; FP = fpr[idx] * N; TN = N - FP
print("")
print("Threshold Value=", thresh[idx])
print("TP=", TP/totalPts, 'FP=', FP/totalPts)
print("FN=", FN/totalPts, "TN=", TN/totalPts)
# 50th
idx = idx50
TP = tpr[idx] * P; FN = P - TP; FP = fpr[idx] * N; TN = N - FP
print("")
print("Threshold Value=", thresh[idx])
print("TP=", TP/totalPts, 'FP=', FP/totalPts)
print("FN=", FN/totalPts, "TN=", TN/totalPts)
# 75th
idx = idx75
TP = tpr[idx] * P; FN = P - TP; FP = fpr[idx] * N; TN = N - FP
print("")
print("Threshold Value=", thresh[idx])
print("TP=", TP/totalPts, 'FP=', FP/totalPts)
print("FN=", FN/totalPts, "TN=", TN/totalPts)7.5 用Python集成方法解决多类别分类问题
用随机森林对玻璃进行分类:代码生成随机森林模型,然后绘制训练过程、属性的重要性排名。打印输出混淆矩阵,此矩阵显示了对于每一个类别,其样本分别有多少被预测成了其他类别。如果分类器是完美的,则在矩阵里不应该有偏离对角线的元素。
__author__ = 'mike-bowles'
import urllib.request as urllib2
import numpy
import matplotlib.pyplot as plot
from sklearn.cross_validation import train_test_split
from sklearn import ensemble
from sklearn.metrics import mean_squared_error, accuracy_score, confusion_matrix, roc_curve
import csv
from io import StringIO as pyIO
target_url = ("http://archive.ics.uci.edu/ml/machine-learning-databases/abalone/abalone.data")
data = urllib2.urlopen(target_url)
data = data.read().decode('utf-8')
dataFile = pyIO(data)
csvReader = csv.reader(dataFile)
xList = []
for line in csvReader:
#print(line)
row = line
#labels.append(float(row.pop()))
xList.append(row)
glassNames = numpy.array(['RI', 'Na', 'Mg', 'Al', 'Si', 'K', 'Ca', 'Ba', 'Fe', 'Type'])
xNum = []
labels = []
for row in xList:
labels.append(row.pop())
l = len(row)
attrRow = [float(row[i]) for i in range(1, l)]
xNum.append(attrRow)
nrows = len(xNum)
ncols = len(xNum[1])
newLabels = []
labelSet = set(labels) # !!!
labelList = list(labelSet)
labelList.sort()
nlabels = len(labelList)
print(labelList)
for l in labels:
index = labelList.index(l)
newLabels.append(index)
print(set(newLabels))
xTemp = [xNum[i] for i in range(nrows) if newLabels[i]==1]
print(xTemp[0])
yTemp = [newLabels[i] for i in range(nrows) if newLabels[i]==1]
xTrain, xTest, yTrain, yTest = train_test_split(xTemp, yTemp, test_size=0.30, random_state=531)
print(xTrain[0])
for iLabel in range(2, len(labelList)):
# segregate x and y according to labels
xTemp = [xNum[i] for i in range(nrows) if newLabels[i]==iLabel]
yTemp = [newLabels[i] for i in range(nrows) if newLabels[i]==iLabel]
#print(len(xTemp[1]))
xTrainTemp, xTestTemp, yTrainTemp, yTestTemp = train_test_split(xTemp, yTemp, test_size=0.30, random_state=531)
#accumulate
if len(xTrainTemp)==0 or len(xTestTemp)==0 or len(yTrainTemp)==0 or len(yTestTemp)==0:
print(iLabel)
continue
xTrain = numpy.append(xTrain, xTrainTemp, axis=0)
xTest = numpy.append(xTest, xTestTemp, axis=0)
yTrain = numpy.append(yTrain, yTrainTemp, axis=0)
yTest = numpy.append(yTest, yTestTemp, axis=0)
print(yTrain)
missClassError = []
nTreeList = range(50, 2000, 50)
for iTrees in nTreeList:
depth = None
maxFeat = 4 # try tweaking
glassRFModel = ensemble.RandomForestClassifier(n_estimators=iTrees, max_depth=depth, max_features=maxFeat, oob_score=False, random_state=531)
glassRFModel.fit(xTrain, yTrain)
# Accumulate auc on test set
prediction = glassRFModel.predict(xTest)
correct = accuracy_score(yTest, prediction)
missClassError.append(1.0 - correct)
print("Missclassification Error")
print(missClassError[-1])
# generate confusion matrix
pList = prediction.tolist()
confusionMat = confusion_matrix(yTest, pList)
print("")
print("Confusion Matrix")
print(confusionMat)
# plot
plot.plot(nTreeList, missClassError)
plot.xlabel("Number of Trees in Ensemble")
plot.ylabel("Missclassification Error Rate")
plot.show()
#
featureImportance = glassRFModel.feature_importances_
featureImportance = featureImportance/featureImportance.max()
sortedIdx = numpy.argsort(featureImportance)
barPos = numpy.arange(sortedIdx.shape[0]) + 0.5
plot.barh(barPos, featureImportance[sortedIdx], align="center")
plot.yticks(barPos, glassNames[sortedIdx])
plot.xlabel('Variable Importance')
plot.subplots_adjust(left=0.2, right=0.9, top=0.9, bottom=0.1)
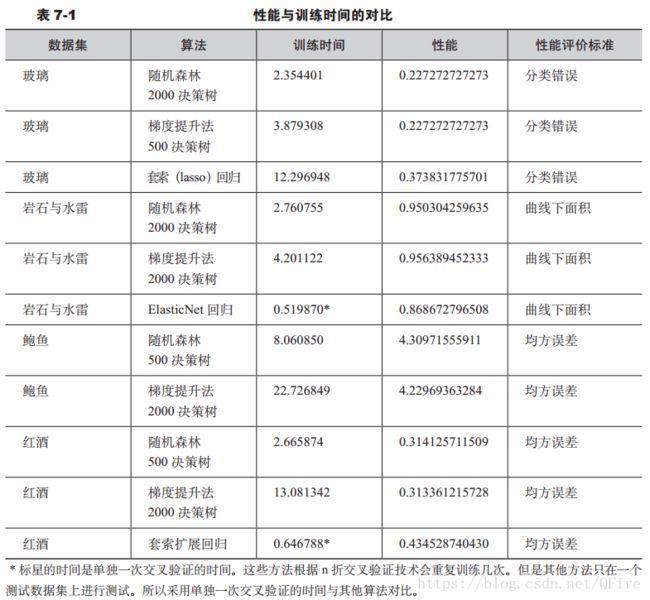
plot.show()7.6 算法比较
随机森林和梯度提升法两者的性能很接近。只不过某些时候,一个算法要比另一个算法需要训练更多的决策树才能达到相似的性能。