数据科学 案例13 聚类与K-mean聚类(代码)
- 15 聚类与K-mean聚类
- 15.1 层次聚类
- 1、手动测试主成分数量
- 2、根据主成分分析确定需要保留的主成分数量,进行因子分析
- 3、根据因子得分进行数据分析
- 15.2 K-means聚类分析
- 15.2.1 测试主成分数量、因子分析
- 15.2.1、使用因子得分进行k-means聚类
- 1、k-means聚类的第一种方式:不进行变量分布的正态转换--用于寻找异常值
- 2、k-means聚类的第二种方式:进行变量分布的正态转换--用于客户细分
15 聚类与K-mean聚类
import os
import pandas as pd
15.1 层次聚类
1、手动测试主成分数量
model_data = pd.read_csv(r'.\data\cities_10.csv',encoding='gbk')
model_data.head()
|
AREA |
X1 |
X2 |
X3 |
X4 |
X5 |
X6 |
X7 |
X8 |
X9 |
| 0 |
辽宁 |
5458.2 |
13000 |
1376.2 |
2258.4 |
1315.9 |
529.0 |
2258.4 |
123.7 |
399.7 |
| 1 |
山东 |
10550.0 |
11643 |
3502.5 |
3851.0 |
2288.7 |
1070.7 |
3181.9 |
211.1 |
610.2 |
| 2 |
河北 |
6076.6 |
9047 |
1406.7 |
2092.6 |
1161.6 |
597.1 |
1968.3 |
45.9 |
302.3 |
| 3 |
天津 |
2022.6 |
22068 |
822.8 |
960.0 |
703.7 |
361.9 |
941.4 |
115.7 |
171.8 |
| 4 |
江苏 |
10636.3 |
14397 |
3536.3 |
3967.2 |
2320.0 |
1141.3 |
3215.8 |
384.7 |
643.7 |
data = model_data.loc[ :,'X1':]
data.head()
|
X1 |
X2 |
X3 |
X4 |
X5 |
X6 |
X7 |
X8 |
X9 |
| 0 |
5458.2 |
13000 |
1376.2 |
2258.4 |
1315.9 |
529.0 |
2258.4 |
123.7 |
399.7 |
| 1 |
10550.0 |
11643 |
3502.5 |
3851.0 |
2288.7 |
1070.7 |
3181.9 |
211.1 |
610.2 |
| 2 |
6076.6 |
9047 |
1406.7 |
2092.6 |
1161.6 |
597.1 |
1968.3 |
45.9 |
302.3 |
| 3 |
2022.6 |
22068 |
822.8 |
960.0 |
703.7 |
361.9 |
941.4 |
115.7 |
171.8 |
| 4 |
10636.3 |
14397 |
3536.3 |
3967.2 |
2320.0 |
1141.3 |
3215.8 |
384.7 |
643.7 |
corr_matrix = data.corr(method='pearson')
corr_matrix
|
X1 |
X2 |
X3 |
X4 |
X5 |
X6 |
X7 |
X8 |
X9 |
| X1 |
1.000000 |
-0.094292 |
0.966506 |
0.979238 |
0.922984 |
0.921680 |
0.941148 |
0.637458 |
0.825568 |
| X2 |
-0.094292 |
1.000000 |
0.112726 |
0.074167 |
0.214052 |
0.093483 |
-0.042776 |
0.081195 |
0.273145 |
| X3 |
0.966506 |
0.112726 |
1.000000 |
0.985373 |
0.963159 |
0.939194 |
0.935196 |
0.704714 |
0.898016 |
| X4 |
0.979238 |
0.074167 |
0.985373 |
1.000000 |
0.972862 |
0.939720 |
0.962267 |
0.713890 |
0.913364 |
| X5 |
0.922984 |
0.214052 |
0.963159 |
0.972862 |
1.000000 |
0.971337 |
0.937109 |
0.716722 |
0.934549 |
| X6 |
0.921680 |
0.093483 |
0.939194 |
0.939720 |
0.971337 |
1.000000 |
0.897127 |
0.624294 |
0.848004 |
| X7 |
0.941148 |
-0.042776 |
0.935196 |
0.962267 |
0.937109 |
0.897127 |
1.000000 |
0.836272 |
0.928692 |
| X8 |
0.637458 |
0.081195 |
0.704714 |
0.713890 |
0.716722 |
0.624294 |
0.836272 |
1.000000 |
0.881528 |
| X9 |
0.825568 |
0.273145 |
0.898016 |
0.913364 |
0.934549 |
0.848004 |
0.928692 |
0.881528 |
1.000000 |
from sklearn import preprocessing
data = preprocessing.scale(data)
from sklearn.decomposition import PCA
'''说明:1、第一次的n_components参数应该设的大一点
说明:2、观察explained_variance_ratio_和explained_variance_的取值变化,建议explained_variance_ratio_累积大于0.85,explained_variance_需要保留的最后一个主成分大于0.8,
'''
pca=PCA(n_components=3)
newData=pca.fit(data)
print(pca.explained_variance_)
print(pca.explained_variance_ratio_)
[8.01129553 1.22149318 0.60792399]
[0.80112955 0.12214932 0.0607924 ]
2、根据主成分分析确定需要保留的主成分数量,进行因子分析
from fa_kit import FactorAnalysis
from fa_kit import plotting as fa_plotting
fa = FactorAnalysis.load_data_samples(
data,
preproc_demean=True,
preproc_scale=True
)
fa.extract_components()
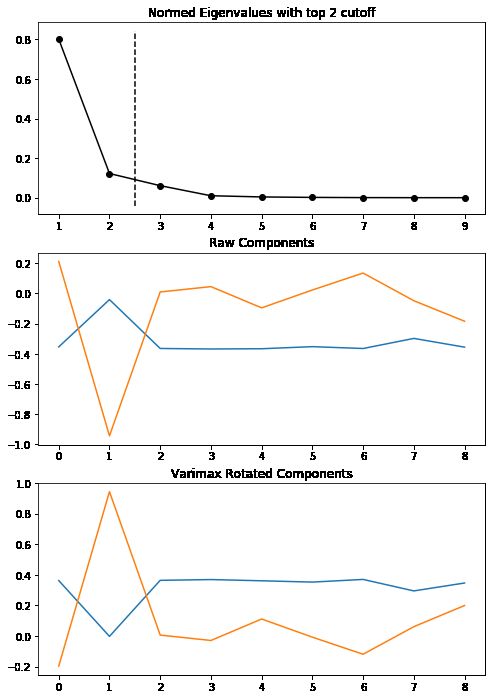
fa.find_comps_to_retain(method='top_n',num_keep=2)
array([0, 1], dtype=int64)
fa.rotate_components(method='varimax')
fa_plotting.graph_summary(fa)
pd.DataFrame(fa.comps["rot"])
|
0 |
1 |
| 0 |
0.362880 |
-0.196047 |
| 1 |
-0.001947 |
0.943648 |
| 2 |
0.364222 |
0.006565 |
| 3 |
0.369255 |
-0.028775 |
| 4 |
0.361258 |
0.111596 |
| 5 |
0.352799 |
-0.007144 |
| 6 |
0.370140 |
-0.118691 |
| 7 |
0.295099 |
0.061400 |
| 8 |
0.346765 |
0.199650 |
import numpy as np
fas = pd.DataFrame(fa.comps["rot"])
data = pd.DataFrame(data)
score = pd.DataFrame(np.dot(data, fas))
score
|
0 |
1 |
| 0 |
-1.174241 |
-0.364178 |
| 1 |
2.095775 |
-0.654819 |
| 2 |
-1.399899 |
-0.870629 |
| 3 |
-3.265185 |
0.698849 |
| 4 |
2.386557 |
-0.337666 |
| 5 |
0.163901 |
2.802894 |
| 6 |
1.209012 |
0.048116 |
| 7 |
-2.084500 |
-0.322173 |
| 8 |
5.501759 |
0.105138 |
| 9 |
-3.433179 |
-1.105531 |
3、根据因子得分进行数据分析
a=score.rename(columns={0: "Gross", 1: "Avg"})
citi10_fa=model_data.join(a)
citi10_fa
|
AREA |
X1 |
X2 |
X3 |
X4 |
X5 |
X6 |
X7 |
X8 |
X9 |
Gross |
Avg |
| 0 |
辽宁 |
5458.2 |
13000 |
1376.2 |
2258.4 |
1315.9 |
529.0 |
2258.4 |
123.7 |
399.7 |
-1.174241 |
-0.364178 |
| 1 |
山东 |
10550.0 |
11643 |
3502.5 |
3851.0 |
2288.7 |
1070.7 |
3181.9 |
211.1 |
610.2 |
2.095775 |
-0.654819 |
| 2 |
河北 |
6076.6 |
9047 |
1406.7 |
2092.6 |
1161.6 |
597.1 |
1968.3 |
45.9 |
302.3 |
-1.399899 |
-0.870629 |
| 3 |
天津 |
2022.6 |
22068 |
822.8 |
960.0 |
703.7 |
361.9 |
941.4 |
115.7 |
171.8 |
-3.265185 |
0.698849 |
| 4 |
江苏 |
10636.3 |
14397 |
3536.3 |
3967.2 |
2320.0 |
1141.3 |
3215.8 |
384.7 |
643.7 |
2.386557 |
-0.337666 |
| 5 |
上海 |
5408.8 |
40627 |
2196.2 |
2755.8 |
1970.2 |
779.3 |
2035.2 |
320.5 |
709.0 |
0.163901 |
2.802894 |
| 6 |
浙江 |
7670.0 |
16570 |
2356.5 |
3065.0 |
2296.6 |
1180.6 |
2877.5 |
294.2 |
566.9 |
1.209012 |
0.048116 |
| 7 |
福建 |
4682.0 |
13510 |
1047.1 |
1859.0 |
964.5 |
397.9 |
1663.3 |
173.7 |
272.9 |
-2.084500 |
-0.322173 |
| 8 |
广东 |
11769.7 |
15030 |
4224.6 |
4793.6 |
3022.9 |
1275.5 |
5013.6 |
1843.7 |
1201.6 |
5.501759 |
0.105138 |
| 9 |
广西 |
2455.4 |
5062 |
367.0 |
995.7 |
542.2 |
352.7 |
1025.5 |
15.1 |
186.7 |
-3.433179 |
-1.105531 |
citi10_fa.to_csv("citi10_fa.csv")
from pylab import mpl
mpl.rcParams['font.sans-serif'] = ['SimHei']
mpl.rcParams['axes.unicode_minus'] = False
import matplotlib.pyplot as plt
x=citi10_fa['Gross']
y=citi10_fa['Avg']
label=citi10_fa['AREA']
plt.scatter(x, y)
for a,b,l in zip(x,y,label):
plt.text(a, b+0.1, '%s.' % l, ha='center', va= 'bottom',fontsize=14)
plt.show()
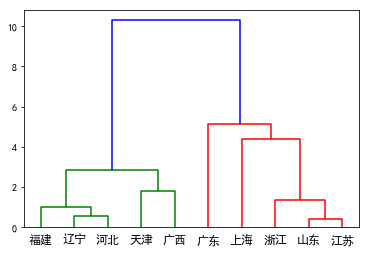
import scipy.cluster.hierarchy as sch
disMat = sch.distance.pdist(citi10_fa[['Gross','Avg']],'euclidean')
Z=sch.linkage(disMat,method='ward')
P=sch.dendrogram(Z,labels=['辽宁','山东','河北','天津','江苏','上海','浙江','福建','广东','广西'])
plt.savefig('plot_dendrogram1.png')
15.2 K-means聚类分析
15.2.1 测试主成分数量、因子分析
import pandas as pd
model_data = pd.read_csv(r".\data\profile_bank.csv")
data = model_data.loc[ :,'CNT_TBM':'CNT_CSC']
data.head()
|
CNT_TBM |
CNT_ATM |
CNT_POS |
CNT_CSC |
| 0 |
34 |
3 |
3 |
9 |
| 1 |
44 |
17 |
5 |
18 |
| 2 |
122 |
26 |
32 |
36 |
| 3 |
42 |
3 |
6 |
1 |
| 4 |
20 |
15 |
2 |
2 |
corr_matrix = data.corr(method='pearson')
corr_matrix
|
CNT_TBM |
CNT_ATM |
CNT_POS |
CNT_CSC |
| CNT_TBM |
1.000000 |
0.055648 |
0.083624 |
0.198835 |
| CNT_ATM |
0.055648 |
1.000000 |
0.341161 |
0.242106 |
| CNT_POS |
0.083624 |
0.341161 |
1.000000 |
0.234055 |
| CNT_CSC |
0.198835 |
0.242106 |
0.234055 |
1.000000 |
from sklearn import preprocessing
data = preprocessing.scale(data)
from sklearn.decomposition import PCA
'''说明:1、第一次的n_components参数应该设的大一点
说明:2、观察explained_variance_ratio_和explained_variance_的取值变化,建议explained_variance_ratio_累积大于0.85,explained_variance_需要保留的最后一个主成分大于0.8,
'''
pca=PCA(n_components=4)
newData=pca.fit(data)
print(pca.explained_variance_)
print(pca.explained_variance_ratio_)
[1.60786876 1.00252275 0.7339482 0.65570029]
[0.40196317 0.25062818 0.18348521 0.16392343]
'''通过主成分在每个变量上的权重的绝对值大小,确定每个主成分的代表性
'''
pd.DataFrame(pca.components_).T
|
0 |
1 |
2 |
3 |
| 0 |
0.303020 |
0.834245 |
0.445132 |
0.118622 |
| 1 |
0.555131 |
-0.377566 |
0.135542 |
0.728630 |
| 2 |
0.559520 |
-0.315486 |
0.386716 |
-0.661708 |
| 3 |
0.535673 |
0.248894 |
-0.796201 |
-0.131035 |
from fa_kit import FactorAnalysis
from fa_kit import plotting as fa_plotting
fa = FactorAnalysis.load_data_samples(
data,
preproc_demean=True,
preproc_scale=True
)
fa.extract_components()
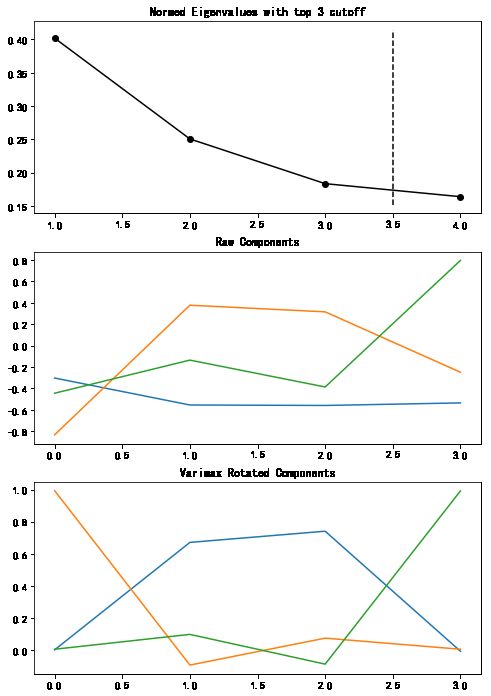
fa.find_comps_to_retain(method='top_n',num_keep=3)
array([0, 1, 2], dtype=int64)
fa.rotate_components(method='varimax')
fa_plotting.graph_summary(fa)
pd.DataFrame(fa.comps["rot"])
import numpy as np
fas = pd.DataFrame(fa.comps["rot"])
data = pd.DataFrame(data)
score = pd.DataFrame(np.dot(data, fas))
fa_scores=score.rename(columns={0: "ATM_POS", 1: "TBM", 2: "CSC"})
fa_scores.head()
|
ATM_POS |
TBM |
CSC |
| 0 |
-0.852354 |
-0.294938 |
0.143935 |
| 1 |
-0.333078 |
-0.244334 |
0.939343 |
| 2 |
0.918067 |
0.593787 |
2.349496 |
| 3 |
-0.741847 |
-0.210507 |
-0.521592 |
| 4 |
-0.499703 |
-0.492714 |
-0.367629 |
15.2.1、使用因子得分进行k-means聚类
1、k-means聚类的第一种方式:不进行变量分布的正态转换–用于寻找异常值
var = ["ATM_POS","TBM","CSC"]
skew_var = {}
for i in var:
skew_var[i]=abs(fa_scores[i].skew())
skew=pd.Series(skew_var).sort_values(ascending=False)
skew
TBM 51.881233
CSC 6.093417
ATM_POS 2.097633
dtype: float64
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=3)
result=kmeans.fit(fa_scores)
result
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
model_data_l=model_data.join(pd.DataFrame(result.labels_))
model_data_l=model_data_l.rename(columns={0: "clustor"})
model_data_l.head()
|
ID |
CNT_TBM |
CNT_ATM |
CNT_POS |
CNT_CSC |
CNT_TOT |
clustor |
| 0 |
41360 |
34 |
3 |
3 |
9 |
49 |
0 |
| 1 |
52094 |
44 |
17 |
5 |
18 |
84 |
0 |
| 2 |
57340 |
122 |
26 |
32 |
36 |
216 |
1 |
| 3 |
76885 |
42 |
3 |
6 |
1 |
52 |
0 |
| 4 |
89150 |
20 |
15 |
2 |
2 |
39 |
0 |
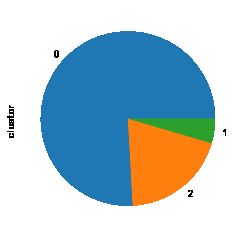
import matplotlib
get_ipython().magic('matplotlib inline')
model_data_l.clustor.value_counts().plot(kind = 'pie')
2、k-means聚类的第二种方式:进行变量分布的正态转换–用于客户细分
import numpy as np
from sklearn import preprocessing
quantile_transformer = preprocessing.QuantileTransformer(output_distribution='normal', random_state=0)
fa_scores_trans=quantile_transformer.fit_transform(fa_scores)
fa_scores_trans=pd.DataFrame(fa_scores_trans)
fa_scores_trans=fa_scores_trans.rename(columns={0: "ATM_POS", 1: "TBM", 2: "CSC"})
fa_scores_trans.head()
|
ATM_POS |
TBM |
CSC |
| 0 |
-0.501859 |
-0.265036 |
0.770485 |
| 1 |
0.097673 |
-0.154031 |
1.316637 |
| 2 |
0.952085 |
1.168354 |
1.845934 |
| 3 |
-0.333179 |
-0.084688 |
-1.780166 |
| 4 |
-0.071278 |
-0.888898 |
-0.066404 |
var = ["ATM_POS","TBM","CSC"]
skew_var = {}
for i in var:
skew_var[i]=abs(fa_scores_trans[i].skew())
skew=pd.Series(skew_var).sort_values(ascending=False)
skew
CSC 5.400815e-04
ATM_POS 4.822917e-04
TBM 2.196784e-07
dtype: float64
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=4)
result=kmeans.fit(fa_scores_trans)
print(result)
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=4, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
model_data_l=model_data.join(pd.DataFrame(result.labels_))
model_data_l=model_data_l.rename(columns={0: "clustor"})
model_data_l.head()
|
ID |
CNT_TBM |
CNT_ATM |
CNT_POS |
CNT_CSC |
CNT_TOT |
clustor |
| 0 |
41360 |
34 |
3 |
3 |
9 |
49 |
3 |
| 1 |
52094 |
44 |
17 |
5 |
18 |
84 |
0 |
| 2 |
57340 |
122 |
26 |
32 |
36 |
216 |
0 |
| 3 |
76885 |
42 |
3 |
6 |
1 |
52 |
1 |
| 4 |
89150 |
20 |
15 |
2 |
2 |
39 |
3 |
import matplotlib
get_ipython().magic('matplotlib inline')
model_data_l.clustor.value_counts().plot(kind = 'pie')
data1 = model_data.loc[ :,'CNT_TBM':'CNT_CSC']
from sklearn import tree
clf = tree.DecisionTreeClassifier(criterion='gini', max_depth=2, min_samples_split=100, min_samples_leaf=100, random_state=12345)
clf.fit(data1, result.labels_)
DecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',
max_depth=2, max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=100, min_samples_split=100,
min_weight_fraction_leaf=0.0, presort='deprecated',
random_state=12345, splitter='best')
import pydotplus
from IPython.display import Image
import sklearn.tree as tree
dot_data = tree.export_graphviz(clf,
out_file=None,
feature_names=data1.columns,
class_names=['0','1','2','3'],
filled=True)
graph = pydotplus.graph_from_dot_data(dot_data)
Image(graph.create_png())