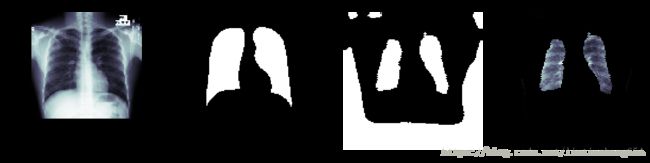kaggle 入门系列翻译(五) RSNA 肺炎预测
第二课:肺部X光结果的语义分割
本课主要介绍应用MD.ai使用U-Net来进行语义分割;
使用MD.ai注释器查看DICOM图像,并创建图像级别注释。然后使用MD.ai python客户端库下载图像和注释,准备数据集,然后用于训练模型进行分类。
MD.ai官网如下:https://www.md.ai/
是一个专门用于医疗AI的开源库,不过目前好像就只有这个kaggle项目呀,可以很方便的看这次比赛里面的各个图片。。
参照第一课安装和导入需要的库并进行初始准备:
这里需要导入一个token,是mdai需要的,进入https://public.md.ai/hub/settings#tokens 页面,登录后左侧有个usersetting 的tab,点进去后可以点生成token。
p = mdai_client.project('aGq4k6NW', path='')
该行代码用于在网上搜索为aGq4k6NW的项目,path表示下载路径,置空表示当前路径。
pip install pydicom
pip install tqdm
pip install imgaug
pip install mdai
import os
import sys
import random
import math
import numpy as np
import cv2
import matplotlib.pyplot as plt
import json
import pydicom
from imgaug import augmenters as iaa
import skimage.io
import skimage.measure
from tqdm import tqdm
from PIL import Image
import requests
import shutil
import zipfile
import mdai
mdai_client = mdai.Client(domain='public.md.ai', access_token="")
p = mdai_client.project('aGq4k6NW', path='./lesson2-data')
p.show_label_groups()
labels_dict = {'L_A8Jm3d':1 # Lung
}
print(labels_dict)
p.set_labels_dict(labels_dict)
p.show_datasets()
dataset = p.get_dataset_by_id('D_rQLwzo')
dataset.prepare()
image_ids = dataset.get_image_ids()
len(image_ids)
# 查看部分训练图片
mdai.visualize.display_images(image_ids[:3], cols=2)下载UNet实现:
简要介绍一下UNet:
UNET_URL = 'https://s3.amazonaws.com/md.ai-ml-lessons/unet.zip'
UNET_ZIPPED = 'unet.zip'
if not os.path.exists(UNET_ZIPPED):
r = requests.get(UNET_URL, stream=True)
if r.status_code == requests.codes.ok:
with open(UNET_ZIPPED, "wb") as f:
shutil.copyfileobj(r.raw, f)
else:
r.raise_for_status()
with zipfile.ZipFile(UNET_ZIPPED) as zf:
zf.extractall()进行训练
imgs_anns_dict = dataset.imgs_anns_dict
from unet import dataset
from unet import dilated_unet
from unet import train
images, masks = dataset.load_images(imgs_anns_dict)
img_index = random.choice(range(len(imgs_anns_dict)))
print(img_index)
img_fps = list(imgs_anns_dict.keys())
img_fp = img_fps[img_index]
img = mdai.visualize.load_dicom_image(img_fp)
ann = imgs_anns_dict[img_fp]
img_width = img.shape[1]
img_height = img.shape[0]
mask = np.zeros((img_height, img_width), dtype=np.uint8)
for a in ann:
vertices = np.array(a['data']['vertices'])
vertices = vertices.reshape((-1,2))
cv2.fillPoly(mask, np.int32([vertices]), (255,255,255))
plt.figure(figsize=(30, 20))
plt.subplot(2,3,1)
plt.imshow(img, cmap=plt.cm.bone)
plt.axis('off')
plt.subplot(2,3,2)
plt.imshow(mask, cmap=plt.cm.bone)
plt.axis('off')
plt.subplot(2,3,3)
plt.imshow(cv2.bitwise_and(img, img, mask=mask.astype(np.uint8)), cmap=plt.cm.bone)
plt.axis('off')
plt.subplot(2,3,4)
plt.imshow(images[img_index,:,:,0], cmap=plt.cm.bone)
plt.axis('off')
plt.subplot(2,3,5)
plt.imshow(masks[img_index,:,:,0], cmap=plt.cm.bone)
plt.axis('off')
plt.subplot(2,3,6)
plt.imshow(cv2.bitwise_and(images[img_index,:,:,0], images[img_index,:,:,0],
mask=masks[img_index,:,:,0].astype(np.uint8)), cmap=plt.cm.bone)
plt.axis('off')
import tensorflow as tf
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
CONFIG_FP = 'unet/configs/11.json'
name = os.path.basename(CONFIG_FP).split('.')[0]
print(name)
with open(CONFIG_FP, 'r') as f:
config = json.load(f)
# increase the number of epochs for better prediction
history = train.train(config, name, images,masks, num_epochs=20)
#画出学习曲线
import matplotlib.pyplot as plt
print(history.history.keys())
plt.figure()
plt.plot(history.history['acc'], 'orange', label='Training accuracy')
plt.plot(history.history['val_acc'], 'blue', label='Validation accuracy')
plt.plot(history.history['loss'], 'red', label='Training loss')
plt.plot(history.history['val_loss'], 'green', label='Validation loss')
plt.legend()
plt.show()
from keras.models import load_model
import keras.backend as K
model_name = 'unet/trained/model_'+name+'.hdf5'
print(model_name)
model = load_model(model_name, custom_objects={'dice': train.dice, 'iou': train.iou})
images, masks = dataset.load_images(imgs_anns_dict)
plt.figure(figsize=(20, 10))
img_index = random.choice(range(len(images)))
plt.subplot(1,4,1)
random_img = images[img_index,:,:,0]
plt.imshow(random_img, cmap=plt.cm.bone)
plt.axis('off')
plt.title('Lung X-Ray')
plt.subplot(1,4,2)
random_mask = masks[img_index,:,:,0]
plt.imshow(random_mask, cmap=plt.cm.bone)
plt.axis('off')
plt.title('Mask Ground Truth')
random_img_2 = np.expand_dims(np.expand_dims(random_img, axis=0), axis=3)
mask = model.predict(random_img_2)[0][:,:,0] > 0.5
plt.subplot(1,4,3)
plt.imshow(mask, cmap=plt.cm.bone)
plt.axis('off')
plt.title('Predicted Mask')
plt.subplot(1,4,4)
plt.imshow(cv2.bitwise_and(random_img, random_img, mask=mask.astype(np.uint8)), cmap=plt.cm.bone)
plt.axis('off')
plt.title('Predicted Lung Segmentation')预测出的肺部区域