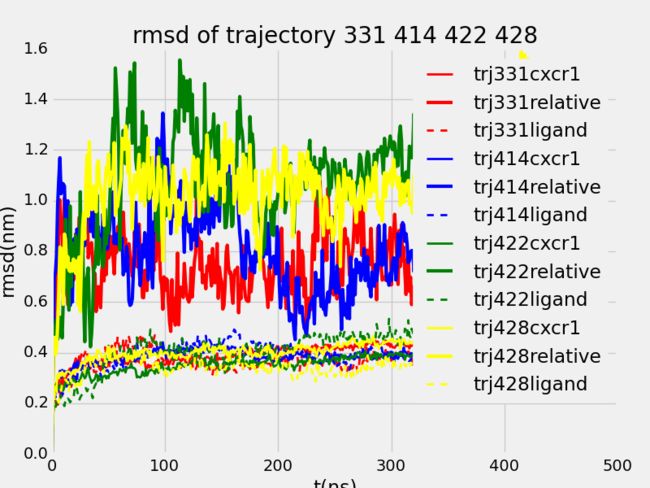
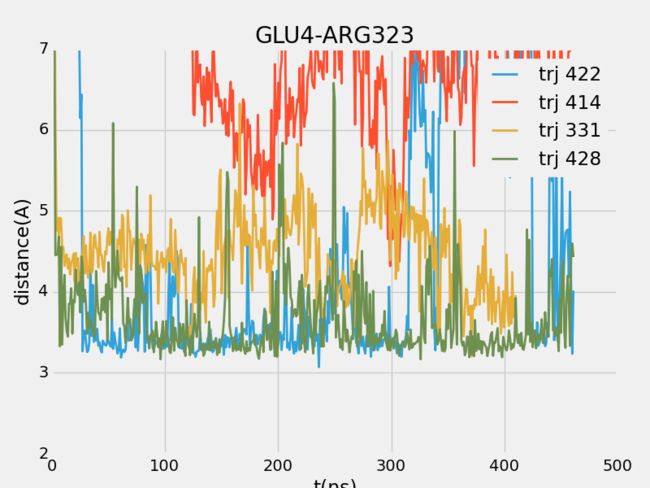
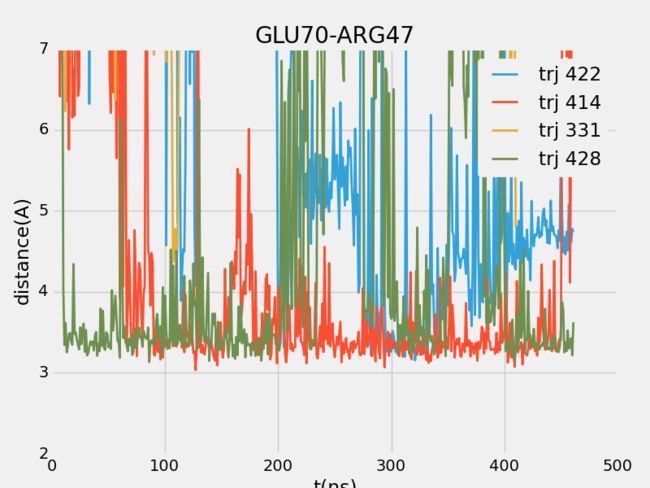
- CesiumJS+SuperMap3D.js混用实现可视域分析 S3M图层加载 裁剪区域绘制
SteveJi666
WebGLcesiumEarthSDKSuperMap3djavascript前端arcgis
版本简介:cesium:1.99;Supermap3D:SuperMapiClientJavaScript11i(2023);官方下载文档链家:SuperMap技术资源中心|为您提供全面的在线技术服务示例参考:support.supermap.com.cn:8090/webgl/Cesium/examples/webgl/examples.html#analysissupport.supermap
- CesiumJS+SuperMap3D.js混用实现通视分析
SteveJi666
WebGLcesiumEarthSDKSuperMap3djavascript前端arcgis
版本简介:cesium:1.99;Supermap3D:SuperMapiClientJavaScript11i(2023);官方下载文档链家:SuperMap技术资源中心|为您提供全面的在线技术服务示例参考:support.supermap.com.cn:8090/webgl/Cesium/examples/webgl/examples.html#analysissupport.supermap
- 概率潜在语义分析(Probabilistic Latent Semantic Analysis,PLSA)—无监督学习方法、概率模型、生成模型、共现模型、非线性模型、参数化模型、批量学习
剑海风云
ArtificialIntelligence人工智能机器学习概率潜在语义分析PLSA
定义输入:设单词集合为W={ω1,ω2,⋯ ,ωM}W=\{\omega_1,\omega_2,\cdots,\omega_M\}W={ω1,ω2,⋯,ωM},文本集合为D={d1,d2,⋯ ,dN}D=\{d_1,d_2,\cdots,d_N\}D={d1,d2,⋯,dN},话题集合为Z={z1,z2,⋯ ,zN}Z=\{z_1,z_2,\cdots,z_N\}Z={z1,z2,⋯,zN},共现
- 算法设计与分析 合并排序的递归实现算法
Jxcupupup
算法算法算法设计与分析
合并排序的递归实现算法。输入:先输入进行合并排序元素的个数,然后依次随机输入(或随机生成)每个数字。输出:元素排序后的结果,数字之间不加任何标识符。示//完整代码在GitHub上//https://github.com/Jxcup/Course_Algorithm_Analysis-Design/blob/main/MergeSort_iteration.cpp//合并排序递归#includeus
- AI学习笔记:pdf-document-layout-analysis
hillstream3
人工智能学习笔记pdfAI编程nlp
一直在学AI,但没有连续的时间来尝试。现在终于失业了,有大把连续的时间来动手。之前准备了一台I5-1400F+RTX360012G的电脑,现在终于派上用场了。由于一直在从事无线通信相关的工作,所以,拿到一份很长的AI可能与通信在哪些方面,能够结合的pdf文档。所以,打算从这份文档开始入手。第一个找到的项目的是这个:https://huggingface.co/HURIDOCS/pdf-docume
- 线性判别分析 (Linear Discriminant Analysis, LDA)
ALGORITHM LOL
人工智能机器学习算法
线性判别分析(LinearDiscriminantAnalysis,LDA)通俗易懂算法线性判别分析(LinearDiscriminantAnalysis,LDA)是一种用于分类和降维的技术。其主要目的是找到一个线性变换,将数据投影到一个低维空间,使得在这个新空间中,不同类别的数据能够更好地分离。线性判别分析的核心思想LDA的基本思路是最大化类间方差(between-classvariance)与
- 404 error when doing workload anlysis using locust on OpenAI API (GPT.35)
营赢盈英
AI人工智能pythonopenailocust
题意:"使用Locust对OpenAIAPI(GPT-3.5)进行工作负载分析时出现404错误。"问题背景:IamtryingtodosomeworkloadanalysisonOpenAIGPT-3.5-TURBOusinglocust."我正在使用Locust对OpenAIGPT-3.5-TURBO进行一些工作负载分析。"fromlocustimportHttpUser,between,tas
- 打卡第13天:《利用python进行数据分析》学习笔记
且不了了
第7章——数据规整化:清理、转换、合并、重塑数据变换http://nbviewer.jupyter.org/github/qiebuliaoliao/data_analysis_python/blob/master/ch7/20180405.ipynb
- 亦菲喊你来学机器学习(20) --PCA数据降维
方世恩
机器学习人工智能深度学习python算法sklearn
文章目录PCA数据降维一、降维二、优缺点三、参数四、实例应用1.读取文件2.分离特征和目标变量3.使用PCA进行降维4.打印特征所占百分比和具体比例5.PCA降维后的数据6.划分数据集7.训练逻辑回归模型8.评估模型性能总结PCA数据降维主成分分析(PrincipalComponentAnalysis,PCA)是一种常用的数据降维技术,它可以在保留数据集中最重要的特征的同时,减少数据的维度。PCA
- python 自动下载ERA5 netCDF4格式数据 INFO Request is queued
水猪1
python
2024-05-2111:18:46,271INFOWelcometotheCDS2024-05-2111:18:46,289INFOSendingrequesttohttps://cds.climate.copernicus.eu/api/v2/resources/reanalysis-era5-pressure-levels2024-05-2111:18:46,512INFORequestis
- Python的情感词典情感分析和情绪计算
yava_free
python大数据人工智能
一.大连理工中文情感词典情感分析(SentimentAnalysis)和情绪分类(EmotionClassification)都是非常重要的文本挖掘手段。情感分析的基本流程如下图所示,通常包括:自定义爬虫抓取文本信息;使用Jieba工具进行中文分词、词性标注;定义情感词典提取每行文本的情感词;通过情感词构建情感矩阵,并计算情感分数;结果评估,包括将情感分数置于0.5到-0.5之间,并可视化显示。目
- 使用Python和Jieba库进行中文情感分析:从文本预处理到模型训练的完整指南
快撑死的鱼
Python算法精解python人工智能开发语言
使用Python和Jieba库进行中文情感分析:从文本预处理到模型训练的完整指南情感分析(SentimentAnalysis)是自然语言处理(NLP)领域中的一个重要分支,旨在从文本中识别出情绪、态度或意见等主观信息。在中文文本处理中,由于语言特性不同于英语,如何高效、准确地分词和提取关键词成为情感分析的关键步骤之一。在这篇文章中,我们将深入探讨如何使用Python和Jieba库进行中文情感分析,
- pytorch计算网络参数量和Flops
Mr_Lowbee
PyTorchpytorch深度学习人工智能
fromtorchsummaryimportsummarysummary(net,input_size=(3,256,256),batch_size=-1)输出的参数是除以一百万(/1000000)M,fromfvcore.nnimportFlopCountAnalysisinputs=torch.randn(1,3,256,256).cuda()flop_counter=FlopCountAna
- 股票中的情侣——配对交易
鸿鹄Max
什么是配对交易?配对交易(PairsTrading)是指八十年代中期华尔街著名投行MorganStanley的数量交易员NunzioTartaglia成立的一个数量分析团队提出的一种市场中性投资策略,,其成员主要是物理学家、数学家、以及计算机学家。GanapathyVidyamurthy在《PairsTrading:QuantitativeMethodsandAnalysis》一书中定义配对交易为
- 多模态大模型论文总结
sudun_03
语言模型算法人工智能
MM1:Methods,Analysis&InsightsfromMultimodalLLMPre-training在这项工作中,我们讨论了建立高性能的多模态大型语言模型(MLLMs)。特别是,我们研究了各种模型结构组件和数据选择的重要性。通过对图像编码器、视觉语言连接器和各种预训练数据选择的仔细而全面的验证,我们确定了几个关键的设计教训。例如,我们证明,与其他已发表的多模式预训练结果相比,对于使
- 科研绘图系列:R语言富集散点图(enrichment scatter plot)
生信学习者1
SCI科研绘图系列r语言数据可视化
介绍富集通路散点图(EnrichmentPathwayScatterPlot)是一种数据可视化工具,用于展示基因集富集分析(GeneSetEnrichmentAnalysis,GSEA)的结果。横坐标是对应基因名称,纵坐标是通路名称,图中的点表示该基因在某个通路下的qvalue,可以简单理解为不同环境下的贡献大小。加载R包导入所需要的R包,在导入前需要用户自己安装。library(readxl)l
- CS269I:Incentives in Computer Science 学习笔记 Lecture 16: Revenue-Maximizing Auctions(收入最大化拍卖)
ldc1513
学习笔记算法博弈论
Lecture16:Revenue-MaximizingAuctions(收入最大化拍卖)1RevenueMaximizationandBayesianAnalysis一直以来,我们关注的都是最大化社会福利的拍卖设计(至少在那些真实出价的场景中)。福利最大化确实是在很多场景中我们最多考虑的事情,比如我们之前看了很长时间的赞助搜索和在线广告。在福利最大化拍卖中,收入也被考虑过,但也仅仅是机制的一个副
- es安装ik分词器
abments
ESelasticsearchjenkins大数据
下载分词器首先确定es对应的版本(假设版本是7.10.0)根据版本下载指定的分词器开始安装在线安装./bin/elasticsearch-plugininstallhttps://github.com/medcl/elasticsearch-analysis-ik/releases/download/v7.10.0/elasticsearch-analysis-ik-7.10.0.zip离线安装-
- 2020-12-05
幸福大黑鸭
IT1.LeetCode:存在重复元素Java编写2020-12-05(217.存在重复元素)2.《Java从入门到精通》明日科技:P331~335阅读记xmind笔记,并自己实现实例。知识点之前确实都学过,但还是再系统复习一下吧。3.《Semantic-awareWorkflowConstructionandAnalysisforDistributedDataAnalyticsSystems》:
- 语音识别 学习笔记2024
AI算法网奇
深度学习基础音视频人工智能
目录dragonfly阿里达摩院FunASR:一款高效的端到端语音识别工具包不错的功能介绍librosa安装语音识别dragonfly阿里达摩院FunASR:一款高效的端到端语音识别工具包不错的功能介绍librosa,一个很有趣的Python库!-简书音频转特征向量GitHub-librosa/librosa:Pythonlibraryforaudioandmusicanalysislibrosa
- Python之Pandas详解
八秒记忆的老男孩
PythonPython基础pythonpandas开发语言
Pandas是Python语言的一个扩展程序库,用于数据分析。Pandas是一个开放源码、BSD许可的库,提供高性能、易于使用的数据结构和数据分析工具。Pandas名字衍生自术语“paneldata”(面板数据)和“Pythondataanalysis”(Python数据分析)。Pandas一个强大的分析结构化数据的工具集,基础是NumPy(提供高性能的矩阵运算)。Pandas可以从各种文件格式比
- 数值分析——LU分解(LU Factorization)
怀帝阍而不见
计算数学c++
本系列整理自博主21年秋季学期本科课程数值分析I的编程作业,内容相对基础,参考书:DavidKincaid,WardCheney-NumericalAnalysisMathematicsofScientificComputing(2002,AmericalMathematicalSociety)目录背景LU分解(LU-Factorization)辅助部分Doolittle分解Cholesky分解定
- ATAM
stone_flower_rain
软件测试ATAM
1.基本信息ATAM:ArchitectureTradeoffAnalysisMethod(构架权衡分析方法),它是评价软件构架的一种综合全面的方法。这种方法不仅可以揭示出构架满足特定质量目标的情况,而且(因为它认识到了构架决策会影响多个质量属性)可以使我们更清楚地认识到质量目标之间的联系——即如何权衡诸多质量目标。2.参与人员评估小组该小组是所评估构架的项目外部的小组。它通常由3~5个人组成。在
- 架构权衡分析法ATAM
崔世勋
软件架构
ArchitectureTradeoffAnalysisMethod使用ATAM方法对软件架构进行评估的目标,是理解架构关于软件的质量属性需求决策的结果。ATAM方法不但提示了架构如何满足特定的质量目标,而且还提供了这些质量目标是如何交互的,即它们之间是如何权衡的。最后欢迎大家访问我的个人网站:1024s
- Xilinx Vivado的RTL分析(RTL analysis)、综合(synthesis)和实现
2401_84185145
程序员fpga开发
理论上,FPGA从编程到下载实现预期功能的过程最少仅需要上述7个步骤中的4、5、6和7,即RTL分析、综合、实现和下载。其中的RTL分析、综合、实现的具体含义和区别又是什么?2、RTL分析(RTLanalysis)一般来讲,通常的设计输入都是Verilog、VHDL或者SystemVerilog等硬件描述语言HDL编写的文件,RTL分析这一步就是将HDL语言转化成逻辑电路图的过程。比如HDL语言描
- 矢量数据的空间分析——叠加分析
进击的码农设计师
叠加分析是对不同的数据进行一系列的集合运算,常用于提取要素的空间隐含信息。1.擦除分析:擦除分析是将输入要素中去除掉与擦除要素的多边形相交的部分,将输入要素处于擦除要素外部边界之外的部分输出到新要素类。打开【系统工具箱→AnalysisTools→叠加分析→擦除】工具,设置输入要素和擦除要素。2.相交分析:相交分析是对输入要素做几何交集操作,输入要素可以是各种几何类型要素(点、线、面)的组合。打开
- pp.weekly.statistical_weekly_analysis_chart 统计周报分析图
小二郎_Ejun
URLpp.weekly.statistical_weekly_analysis_chart请求方式POST请求参数参数名类型必填说明token[string]是无skip_type[string]是一级获取查询的类型skip_value[string]是一级获取查询的数据key_value[string]是对应的key,取参类型query_time[string]否查询时间,时间戳秒(默认今天)
- 情感分析相关汇总
宁缺100
自然语言处理自然语言处理情感分析
文章目录情感分析语音情感识别句子or文档级别情感分析情感词汇字典大连理工大学中文情感词汇本体中文金融情感词典金融社交媒体数据应用的市场情绪词典中文情感分析常用词典台湾大学NTUSD简体中文情感词典BosonNLPABSA细腻度情感分析相关比赛【千言情感分析】SKEP句子级情感分析相关博客或者论文中文情感分析(SentimentAnalysis)的难点在哪?现在做得比较好的有哪几家?文本挖掘在商品评
- OHIF Viewer医学影像学习日记
刘斩仙的笔记本
javascriptOHIFViewer医学影像vuereact
前言:OHIFViewer一个开源的,基于Web的,医学影像查看器。项目文档GitHub项目大概流程:我们下载OHIFViewer项目运行打包,发布到服务器,然后暴露访问地址;再由后端提供返回固定格式json的接口,完整路径例如:http://www.baidu.com/#/viewer?url=http://www.your.com/apiv1/dicom/analysis/studies把此链
- 2020-12-17
幸福大黑鸭
IT1.LeetCode:各位相加Java编写2020-12-17(258.各位相加)2.《Java从入门到精通》明日科技:P391~395阅读记xmind笔记,并自己实现实例。知识点之前确实都学过,但还是再系统复习一下吧。3.《Semantic-awareWorkflowConstructionandAnalysisforDistributedDataAnalyticsSystems》:精读关键
- 继之前的线程循环加到窗口中运行
3213213333332132
javathreadJFrameJPanel
之前写了有关java线程的循环执行和结束,因为想制作成exe文件,想把执行的效果加到窗口上,所以就结合了JFrame和JPanel写了这个程序,这里直接贴出代码,在窗口上运行的效果下面有附图。
package thread;
import java.awt.Graphics;
import java.text.SimpleDateFormat;
import java.util
- linux 常用命令
BlueSkator
linux命令
1.grep
相信这个命令可以说是大家最常用的命令之一了。尤其是查询生产环境的日志,这个命令绝对是必不可少的。
但之前总是习惯于使用 (grep -n 关键字 文件名 )查出关键字以及该关键字所在的行数,然后再用 (sed -n '100,200p' 文件名),去查出该关键字之后的日志内容。
但其实还有更简便的办法,就是用(grep -B n、-A n、-C n 关键
- php heredoc原文档和nowdoc语法
dcj3sjt126com
PHPheredocnowdoc
<!doctype html>
<html lang="en">
<head>
<meta charset="utf-8">
<title>Current To-Do List</title>
</head>
<body>
<?
- overflow的属性
周华华
JavaScript
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml&q
- 《我所了解的Java》——总体目录
g21121
java
准备用一年左右时间写一个系列的文章《我所了解的Java》,目录及内容会不断完善及调整。
在编写相关内容时难免出现笔误、代码无法执行、名词理解错误等,请大家及时指出,我会第一时间更正。
&n
- [简单]docx4j常用方法小结
53873039oycg
docx
本代码基于docx4j-3.2.0,在office word 2007上测试通过。代码如下:
import java.io.File;
import java.io.FileInputStream;
import ja
- Spring配置学习
云端月影
spring配置
首先来看一个标准的Spring配置文件 applicationContext.xml
<?xml version="1.0" encoding="UTF-8"?>
<beans xmlns="http://www.springframework.org/schema/beans"
xmlns:xsi=&q
- Java新手入门的30个基本概念三
aijuans
java新手java 入门
17.Java中的每一个类都是从Object类扩展而来的。 18.object类中的equal和toString方法。 equal用于测试一个对象是否同另一个对象相等。 toString返回一个代表该对象的字符串,几乎每一个类都会重载该方法,以便返回当前状态的正确表示.(toString 方法是一个很重要的方法) 19.通用编程:任何类类型的所有值都可以同object类性的变量来代替。
- 《2008 IBM Rational 软件开发高峰论坛会议》小记
antonyup_2006
软件测试敏捷开发项目管理IBM活动
我一直想写些总结,用于交流和备忘,然都没提笔,今以一篇参加活动的感受小记开个头,呵呵!
其实参加《2008 IBM Rational 软件开发高峰论坛会议》是9月4号,那天刚好调休.但接着项目颇为忙,所以今天在中秋佳节的假期里整理了下.
参加这次活动是一个朋友给的一个邀请书,才知道有这样的一个活动,虽然现在项目暂时没用到IBM的解决方案,但觉的参与这样一个活动可以拓宽下视野和相关知识.
- PL/SQL的过程编程,异常,声明变量,PL/SQL块
百合不是茶
PL/SQL的过程编程异常PL/SQL块声明变量
PL/SQL;
过程;
符号;
变量;
PL/SQL块;
输出;
异常;
PL/SQL 是过程语言(Procedural Language)与结构化查询语言(SQL)结合而成的编程语言PL/SQL 是对 SQL 的扩展,sql的执行时每次都要写操作
- Mockito(三)--完整功能介绍
bijian1013
持续集成mockito单元测试
mockito官网:http://code.google.com/p/mockito/,打开documentation可以看到官方最新的文档资料。
一.使用mockito验证行为
//首先要import Mockito
import static org.mockito.Mockito.*;
//mo
- 精通Oracle10编程SQL(8)使用复合数据类型
bijian1013
oracle数据库plsql
/*
*使用复合数据类型
*/
--PL/SQL记录
--定义PL/SQL记录
--自定义PL/SQL记录
DECLARE
TYPE emp_record_type IS RECORD(
name emp.ename%TYPE,
salary emp.sal%TYPE,
dno emp.deptno%TYPE
);
emp_
- 【Linux常用命令一】grep命令
bit1129
Linux常用命令
grep命令格式
grep [option] pattern [file-list]
grep命令用于在指定的文件(一个或者多个,file-list)中查找包含模式串(pattern)的行,[option]用于控制grep命令的查找方式。
pattern可以是普通字符串,也可以是正则表达式,当查找的字符串包含正则表达式字符或者特
- mybatis3入门学习笔记
白糖_
sqlibatisqqjdbc配置管理
MyBatis 的前身就是iBatis,是一个数据持久层(ORM)框架。 MyBatis 是支持普通 SQL 查询,存储过程和高级映射的优秀持久层框架。MyBatis对JDBC进行了一次很浅的封装。
以前也学过iBatis,因为MyBatis是iBatis的升级版本,最初以为改动应该不大,实际结果是MyBatis对配置文件进行了一些大的改动,使整个框架更加方便人性化。
- Linux 命令神器:lsof 入门
ronin47
lsof
lsof是系统管理/安全的尤伯工具。我大多数时候用它来从系统获得与网络连接相关的信息,但那只是这个强大而又鲜为人知的应用的第一步。将这个工具称之为lsof真实名副其实,因为它是指“列出打开文件(lists openfiles)”。而有一点要切记,在Unix中一切(包括网络套接口)都是文件。
有趣的是,lsof也是有着最多
- java实现两个大数相加,可能存在溢出。
bylijinnan
java实现
import java.math.BigInteger;
import java.util.regex.Matcher;
import java.util.regex.Pattern;
public class BigIntegerAddition {
/**
* 题目:java实现两个大数相加,可能存在溢出。
* 如123456789 + 987654321
- Kettle学习资料分享,附大神用Kettle的一套流程完成对整个数据库迁移方法
Kai_Ge
Kettle
Kettle学习资料分享
Kettle 3.2 使用说明书
目录
概述..........................................................................................................................................7
1.Kettle 资源库管
- [货币与金融]钢之炼金术士
comsci
金融
自古以来,都有一些人在从事炼金术的工作.........但是很少有成功的
那么随着人类在理论物理和工程物理上面取得的一些突破性进展......
炼金术这个古老
- Toast原来也可以多样化
dai_lm
androidtoast
Style 1: 默认
Toast def = Toast.makeText(this, "default", Toast.LENGTH_SHORT);
def.show();
Style 2: 顶部显示
Toast top = Toast.makeText(this, "top", Toast.LENGTH_SHORT);
t
- java数据计算的几种解决方法3
datamachine
javahadoopibatisr-languer
4、iBatis
简单敏捷因此强大的数据计算层。和Hibernate不同,它鼓励写SQL,所以学习成本最低。同时它用最小的代价实现了计算脚本和JAVA代码的解耦,只用20%的代价就实现了hibernate 80%的功能,没实现的20%是计算脚本和数据库的解耦。
复杂计算环境是它的弱项,比如:分布式计算、复杂计算、非数据
- 向网页中插入透明Flash的方法和技巧
dcj3sjt126com
htmlWebFlash
将
Flash 作品插入网页的时候,我们有时候会需要将它设为透明,有时候我们需要在Flash的背面插入一些漂亮的图片,搭配出漂亮的效果……下面我们介绍一些将Flash插入网页中的一些透明的设置技巧。
一、Swf透明、无坐标控制 首先教大家最简单的插入Flash的代码,透明,无坐标控制: 注意wmode="transparent"是控制Flash是否透明
- ios UICollectionView的使用
dcj3sjt126com
UICollectionView的使用有两种方法,一种是继承UICollectionViewController,这个Controller会自带一个UICollectionView;另外一种是作为一个视图放在普通的UIViewController里面。
个人更喜欢第二种。下面采用第二种方式简单介绍一下UICollectionView的使用。
1.UIViewController实现委托,代码如
- Eos平台java公共逻辑
蕃薯耀
Eos平台java公共逻辑Eos平台java公共逻辑
Eos平台java公共逻辑
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
蕃薯耀 2015年6月1日 17:20:4
- SpringMVC4零配置--Web上下文配置【MvcConfig】
hanqunfeng
springmvc4
与SpringSecurity的配置类似,spring同样为我们提供了一个实现类WebMvcConfigurationSupport和一个注解@EnableWebMvc以帮助我们减少bean的声明。
applicationContext-MvcConfig.xml
<!-- 启用注解,并定义组件查找规则 ,mvc层只负责扫描@Controller -->
<
- 解决ie和其他浏览器poi下载excel文件名乱码
jackyrong
Excel
使用poi,做传统的excel导出,然后想在浏览器中,让用户选择另存为,保存用户下载的xls文件,这个时候,可能的是在ie下出现乱码(ie,9,10,11),但在firefox,chrome下没乱码,
因此必须综合判断,编写一个工具类:
/**
*
* @Title: pro
- 挥洒泪水的青春
lampcy
编程生活程序员
2015年2月28日,我辞职了,离开了相处一年的触控,转过身--挥洒掉泪水,毅然来到了兄弟连,背负着许多的不解、质疑——”你一个零基础、脑子又不聪明的人,还敢跨行业,选择Unity3D?“,”真是不自量力••••••“,”真是初生牛犊不怕虎•••••“,••••••我只是淡淡一笑,拎着行李----坐上了通向挥洒泪水的青春之地——兄弟连!
这就是我青春的分割线,不后悔,只会去用泪水浇灌——已经来到
- 稳增长之中国股市两点意见-----严控做空,建立涨跌停版停牌重组机制
nannan408
对于股市,我们国家的监管还是有点拼的,但始终拼不过飞流直下的恐慌,为什么呢?
笔者首先支持股市的监管。对于股市越管越荡的现象,笔者认为首先是做空力量超过了股市自身的升力,并且对于跌停停牌重组的快速反应还没建立好,上市公司对于股价下跌没有很好的利好支撑。
我们来看美国和香港是怎么应对股灾的。美国是靠禁止重要股票做空,在
- 动态设置iframe高度(iframe高度自适应)
Rainbow702
JavaScriptiframecontentDocument高度自适应局部刷新
如果需要对画面中的部分区域作局部刷新,大家可能都会想到使用ajax。
但有些情况下,须使用在页面中嵌入一个iframe来作局部刷新。
对于使用iframe的情况,发现有一个问题,就是iframe中的页面的高度可能会很高,但是外面页面并不会被iframe内部页面给撑开,如下面的结构:
<div id="content">
<div id=&quo
- 用Rapael做图表
tntxia
rap
function drawReport(paper,attr,data){
var width = attr.width;
var height = attr.height;
var max = 0;
&nbs
- HTML5 bootstrap2网页兼容(支持IE10以下)
xiaoluode
html5bootstrap
<!DOCTYPE html>
<html>
<head lang="zh-CN">
<meta charset="UTF-8">
<meta http-equiv="X-UA-Compatible" content="IE=edge">