Datacamp 笔记&代码 Supervised Learning with scikit-learn 第四章 Preprocessing and pipelines
更多原始数据文档和JupyterNotebook
Github: https://github.com/JinnyR/Datacamp_DataScienceTrack_Python
Datacamp track: Data Scientist with Python - Course 21 (4)
Exercise
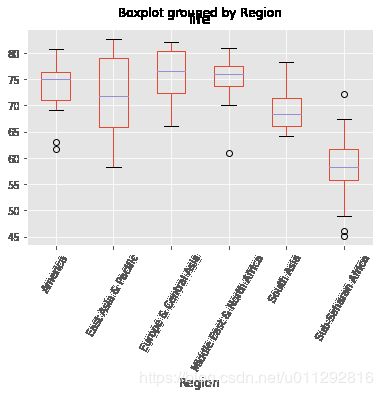
Exploring categorical features
The Gapminder dataset that you worked with in previous chapters also contained a categorical 'Region' feature, which we dropped in previous exercises since you did not have the tools to deal with it. Now however, you do, so we have added it back in!
Your job in this exercise is to explore this feature. Boxplots are particularly useful for visualizing categorical features such as this.
Instruction
- Import
pandasaspd. - Read the CSV file
'gapminder.csv'into a DataFrame calleddf. - Use pandas to create a boxplot showing the variation of life expectancy (
'life') by region ('Region'). To do so, pass the column names in todf.boxplot()(in that order).
import matplotlib.pyplot as plt
plt.style.use('ggplot')
from urllib.request import urlretrieve
fn = 'https://s3.amazonaws.com/assets.datacamp.com/production/course_1939/datasets/gm_2008_region.csv'
urlretrieve(fn, 'gapminder.csv')
('gapminder.csv', )
# Import pandas
import pandas as pd
# Read 'gapminder.csv' into a DataFrame: df
df = pd.read_csv('gapminder.csv')
# Create a boxplot of life expectancy per region
df.boxplot('life', 'Region', rot=60)
# Show the plot
plt.show()
Exercise
Creating dummy variables
As Andy discussed in the video, scikit-learn does not accept non-numerical features. You saw in the previous exercise that the 'Region' feature contains very useful information that can predict life expectancy. For example, Sub-Saharan Africa has a lower life expectancy compared to Europe and Central Asia. Therefore, if you are trying to predict life expectancy, it would be preferable to retain the 'Region' feature. To do this, you need to binarize it by creating dummy variables, which is what you will do in this exercise.
Instruction
- Use the pandas
get_dummies()function to create dummy variables from thedfDataFrame. Store the result asdf_region. - Print the columns of
df_region. This has been done for you. - Use the
get_dummies()function again, this time specifyingdrop_first=Trueto drop the unneeded dummy variable (in this case,'Region_America'). - Hit 'Submit Answer to print the new columns of
df_regionand take note of how one column was dropped!
# Create dummy variables: df_region
df_region = pd.get_dummies(df)
# Print the columns of df_region
print(df_region.columns)
# Create dummy variables with drop_first=True: df_region
df_region = pd.get_dummies(df, drop_first=True)
# Print the new columns of df_region
print(df_region.columns)
Index(['population', 'fertility', 'HIV', 'CO2', 'BMI_male', 'GDP',
'BMI_female', 'life', 'child_mortality', 'Region_America',
'Region_East Asia & Pacific', 'Region_Europe & Central Asia',
'Region_Middle East & North Africa', 'Region_South Asia',
'Region_Sub-Saharan Africa'],
dtype='object')
Index(['population', 'fertility', 'HIV', 'CO2', 'BMI_male', 'GDP',
'BMI_female', 'life', 'child_mortality', 'Region_East Asia & Pacific',
'Region_Europe & Central Asia', 'Region_Middle East & North Africa',
'Region_South Asia', 'Region_Sub-Saharan Africa'],
dtype='object')
Exercise
Regression with categorical features
Having created the dummy variables from the 'Region'feature, you can build regression models as you did before. Here, you’ll use ridge regression to perform 5-fold cross-validation.
The feature array X and target variable array y have been pre-loaded.
Instruction
- Import
Ridgefromsklearn.linear_modelandcross_val_scorefromsklearn.model_selection. - Instantiate a ridge regressor called
ridgewithalpha=0.5andnormalize=True. - Perform 5-fold cross-validation on
Xandyusing thecross_val_score()function. - Print the cross-validated scores.
# modified/added by Jinny
import pandas as pd
import numpy as np
from urllib.request import urlretrieve
fn = 'https://s3.amazonaws.com/assets.datacamp.com/production/course_1939/datasets/gm_2008_region.csv'
urlretrieve(fn, 'gapminder.csv')
df = pd.read_csv('gapminder.csv')
df_region = pd.get_dummies(df)
df_region = df_region.drop('Region_America', axis=1)
X = df_region.drop('life', axis=1).values
y = df_region['life'].values
# Import necessary modules
from sklearn.linear_model import Ridge
from sklearn.model_selection import cross_val_score
# Instantiate a ridge regressor: ridge
ridge = Ridge(alpha=0.5, normalize=True)
# Perform 5-fold cross-validation: ridge_cv
ridge_cv = cross_val_score(ridge, X, y, cv=5)
# Print the cross-validated scores
print(ridge_cv)
[0.86808336 0.80623545 0.84004203 0.7754344 0.87503712]
Exercise
Dropping missing data
The voting dataset from Chapter 1 contained a bunch of missing values that we dealt with for you behind the scenes. Now, it’s time for you to take care of these yourself!
The unprocessed dataset has been loaded into a DataFrame df. Explore it in the IPython Shell with the .head()method. You will see that there are certain data points labeled with a '?'. These denote missing values. As you saw in the video, different datasets encode missing values in different ways. Sometimes it may be a '9999', other times a 0 - real-world data can be very messy! If you’re lucky, the missing values will already be encoded as NaN. We use NaNbecause it is an efficient and simplified way of internally representing missing data, and it lets us take advantage of pandas methods such as .dropna() and .fillna(), as well as scikit-learn’s Imputation transformer Imputer().
In this exercise, your job is to convert the '?'s to NaNs, and then drop the rows that contain them from the DataFrame.
Instruction
- Explore the DataFrame
dfin the IPython Shell. Notice how the missing value is represented. - Convert all
'?'data points tonp.nan. - Count the total number of NaNs using the
.isnull()and.sum()methods. This has been done for you. - Drop the rows with missing values from
dfusing.dropna(). - Hit ‘Submit Answer’ to see how many rows were lost by dropping the missing values.
# modified/added by Jinny
import numpy as np
import pandas as pd
df = pd.read_csv('https://s3.amazonaws.com/assets.datacamp.com/production/course_1939/datasets/house-votes-84.csv',
header=None, names = ['infants', 'water', 'budget', 'physician', 'salvador', 'religious',
'satellite', 'aid', 'missile', 'immigration', 'synfuels', 'education',
'superfund', 'crime', 'duty_free_exports', 'eaa_rsa'])
df = df.reset_index()
df.rename(columns = {'index': 'party'}, inplace = True)
df[df == 'y'] = 1
df[df == 'n'] = 0
# Convert '?' to NaN
df[df == '?'] = np.nan
# Print the number of NaNs
print(df.isnull().sum())
# Print shape of original DataFrame
print("Shape of Original DataFrame: {}".format(df.shape))
# Drop missing values and print shape of new DataFrame
df = df.dropna()
# Print shape of new DataFrame
print("Shape of DataFrame After Dropping All Rows with Missing Values: {}".format(df.shape))
party 0
infants 12
water 48
budget 11
physician 11
salvador 15
religious 11
satellite 14
aid 15
missile 22
immigration 7
synfuels 21
education 31
superfund 25
crime 17
duty_free_exports 28
eaa_rsa 104
dtype: int64
Shape of Original DataFrame: (435, 17)
Shape of DataFrame After Dropping All Rows with Missing Values: (232, 17)
Exercise
Imputing missing data in a ML Pipeline I
As you’ve come to appreciate, there are many steps to building a model, from creating training and test sets, to fitting a classifier or regressor, to tuning its parameters, to evaluating its performance on new data. Imputation can be seen as the first step of this machine learning process, the entirety of which can be viewed within the context of a pipeline. Scikit-learn provides a pipeline constructor that allows you to piece together these steps into one process and thereby simplify your workflow.
You’ll now practice setting up a pipeline with two steps: the imputation step, followed by the instantiation of a classifier. You’ve seen three classifiers in this course so far: k-NN, logistic regression, and the decision tree. You will now be introduced to a fourth one - the Support Vector Machine, or SVM. For now, do not worry about how it works under the hood. It works exactly as you would expect of the scikit-learn estimators that you have worked with previously, in that it has the same .fit() and .predict() methods as before.
Instruction
- Import
Imputerfromsklearn.preprocessingandSVCfromsklearn.svm. SVC stands for Support Vector Classification, which is a type of SVM. - Setup the Imputation transformer to impute missing data (represented as
'NaN') with the'most_frequent'value in the column (axis=0). - Instantiate a
SVCclassifier. Store the result inclf. - Create the steps of the pipeline by creating a list of tuples:
- The first tuple should consist of the imputation step, using
imp. - The second should consist of the classifier.
- The first tuple should consist of the imputation step, using
# Import the Imputer module
from sklearn.preprocessing import Imputer
from sklearn.svm import SVC
# Setup the Imputation transformer: imp
imp = Imputer(missing_values='NaN', strategy='most_frequent', axis=0)
# Instantiate the SVC classifier: clf
clf = SVC()
# Setup the pipeline with the required steps: steps
steps = [('imputation', imp),
('SVM', clf)]
Exercise
Imputing missing data in a ML Pipeline II
Having setup the steps of the pipeline in the previous exercise, you will now use it on the voting dataset to classify a Congressman’s party affiliation. What makes pipelines so incredibly useful is the simple interface that they provide. You can use the .fit() and .predict() methods on pipelines just as you did with your classifiers and regressors!
Practice this for yourself now and generate a classification report of your predictions. The steps of the pipeline have been set up for you, and the feature array X and target variable array y have been pre-loaded. Additionally, train_test_split and classification_report have been imported from sklearn.model_selection and sklearn.metrics respectively.
Instruction
- Import the following modules:
Imputerfromsklearn.preprocessingandPipelinefromsklearn.pipeline.SVCfromsklearn.svm.
- Create the pipeline using
Pipeline()andsteps. - Create training and test sets. Use 30% of the data for testing and a random state of
42. - Fit the pipeline to the training set and predict the labels of the test set.
- Compute the classification report.
# modified/added by Jinny
import pandas as pd
import numpy as np
#
df = pd.read_csv('https://s3.amazonaws.com/assets.datacamp.com/production/course_1939/datasets/votes-ch1.csv')
# Create arrays for the features and the response variable. As a reminder, the response variable is 'party'
y = df['party']
X = df.drop('party', axis=1)
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
# Import necessary modules
from sklearn.preprocessing import Imputer
from sklearn.pipeline import Pipeline
from sklearn.svm import SVC
# Setup the pipeline steps: steps
steps = [('imputation', Imputer(missing_values='NaN', strategy='most_frequent', axis=0)),
('SVM', SVC())]
# Create the pipeline: pipeline
pipeline = Pipeline(steps)
# Create training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Fit the pipeline to the train set
pipeline.fit(X_train, y_train)
# Predict the labels of the test set
y_pred = pipeline.predict(X_test)
# Compute metrics
print(classification_report(y_test, y_pred))
precision recall f1-score support
democrat 0.99 0.96 0.98 85
republican 0.94 0.98 0.96 46
micro avg 0.97 0.97 0.97 131
macro avg 0.96 0.97 0.97 131
weighted avg 0.97 0.97 0.97 131
Exercise
Centering and scaling your data
In the video, Hugo demonstrated how significantly the performance of a model can improve if the features are scaled. Note that this is not always the case: In the Congressional voting records dataset, for example, all of the features are binary. In such a situation, scaling will have minimal impact.
You will now explore scaling for yourself on a new dataset - White Wine Quality! Hugo used the Red Wine Quality dataset in the video. We have used the 'quality' feature of the wine to create a binary target variable: If 'quality'is less than 5, the target variable is 1, and otherwise, it is 0.
The DataFrame has been pre-loaded as df, along with the feature and target variable arrays X and y. Explore it in the IPython Shell. Notice how some features seem to have different units of measurement. 'density', for instance, takes values between 0.98 and 1.04, while 'total sulfur dioxide' ranges from 9 to 440. As a result, it may be worth scaling the features here. Your job in this exercise is to scale the features and compute the mean and standard deviation of the unscaled features compared to the scaled features.
Instruction
- Import
scalefromsklearn.preprocessing. - Scale the features
Xusingscale(). - Print the mean and standard deviation of the unscaled features
X, and then the scaled featuresX_scaled. Use the numpy functionsnp.mean()andnp.std()to compute the mean and standard deviations. - Compute the classification report.
# modified/added by Jinny
import pandas as pd
import numpy as np
df = pd.read_csv('https://s3.amazonaws.com/assets.datacamp.com/production/course_3238/datasets/white-wine.csv')
X = df.drop('quality' , 1).values # drop target variable
y1 = df['quality'].values
y = y1 <= 5
# Import scale
from sklearn.preprocessing import scale
# Scale the features: X_scaled
X_scaled = scale(X)
# Print the mean and standard deviation of the unscaled features
print("Mean of Unscaled Features: {}".format(np.mean(X)))
print("Standard Deviation of Unscaled Features: {}".format(np.std(X)))
# Print the mean and standard deviation of the scaled features
print("Mean of Scaled Features: {}".format(np.mean(X_scaled)))
print("Standard Deviation of Scaled Features: {}".format(np.std(X_scaled)))
Mean of Unscaled Features: 18.432687072460002
Standard Deviation of Unscaled Features: 41.54494764094571
Mean of Scaled Features: 2.7314972981668206e-15
Standard Deviation of Scaled Features: 0.9999999999999999
Exercise
Centering and scaling in a pipeline
With regard to whether or not scaling is effective, the proof is in the pudding! See for yourself whether or not scaling the features of the White Wine Quality dataset has any impact on its performance. You will use a k-NN classifier as part of a pipeline that includes scaling, and for the purposes of comparison, a k-NN classifier trained on the unscaled data has been provided.
The feature array and target variable array have been pre-loaded as X and y. Additionally, KNeighborsClassifier and train_test_split have been imported from sklearn.neighbors and sklearn.model_selection, respectively.
Instruction
- Import the following modules:
StandardScalerfromsklearn.preprocessing.Pipelinefromsklearn.pipeline.
- Complete the steps of the pipeline with
StandardScaler()for'scaler'andKNeighborsClassifier()for'knn'. - Create the pipeline using
Pipeline()andsteps. - Create training and test sets, with 30% used for testing. Use a random state of
42. - Fit the pipeline to the training set.
- Compute the accuracy scores of the scaled and unscaled models by using the
.score()method inside the providedprint()functions.
# modified/added by Jinny
import pandas as pd
df = pd.read_csv('https://s3.amazonaws.com/assets.datacamp.com/production/course_3238/datasets/white-wine.csv')
X = df.drop('quality' , 1).values # drop target variable
y1 = df['quality'].values
y = y1 <= 5
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
# Import the necessary modules
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
# Setup the pipeline steps: steps
steps = [('scaler', StandardScaler()),
('knn', KNeighborsClassifier())]
# Create the pipeline: pipeline
pipeline = Pipeline(steps)
# Create train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Fit the pipeline to the training set: knn_scaled
knn_scaled = pipeline.fit(X_train, y_train)
# Instantiate and fit a k-NN classifier to the unscaled data
knn_unscaled = KNeighborsClassifier().fit(X_train, y_train)
# Compute and print metrics
print('Accuracy with Scaling: {}'.format(knn_scaled.score(X_test, y_test)))
print('Accuracy without Scaling: {}'.format(knn_unscaled.score(X_test, y_test)))
Accuracy with Scaling: 0.7700680272108843
Accuracy without Scaling: 0.6979591836734694
Exercise
Bringing it all together I: Pipeline for classification
It is time now to piece together everything you have learned so far into a pipeline for classification! Your job in this exercise is to build a pipeline that includes scaling and hyperparameter tuning to classify wine quality.
You’ll return to using the SVM classifier you were briefly introduced to earlier in this chapter. The hyperparameters you will tune are C C C and gammagamma. C C C controls the regularization strength. It is analogous to the C C C you tuned for logistic regression in Chapter 3, while gammagamma controls the kernel coefficient: Do not worry about this now as it is beyond the scope of this course.
The following modules have been pre-loaded: Pipeline, svm, train_test_split, GridSearchCV, classification_report, accuracy_score. The feature and target variable arrays X and y have also been pre-loaded.
Instruction
- Setup the pipeline with the following steps:
- Scaling, called
'scaler'withStandardScaler(). - Classification, called
'SVM'withSVC().
- Scaling, called
- Specify the hyperparameter space using the following notation:
'step_name__parameter_name'. Here, thestep_nameisSVM, and theparameter_names areCandgamma. - Create training and test sets, with 20% of the data used for the test set. Use a random state of
21. - Instantiate
GridSearchCVwith the pipeline and hyperparameter space and fit it to the training set. Use 3-fold cross-validation (This is the default, so you don’t have to specify it). - Predict the labels of the test set and compute the metrics. The metrics have been computed for you.
# modified/added by Jinny
import pandas as pd
df = pd.read_csv('https://s3.amazonaws.com/assets.datacamp.com/production/course_3238/datasets/white-wine.csv')
X = df.drop('quality' , 1).values # drop target variable
y1 = df['quality'].values
y = y1 <= 5
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
from sklearn.model_selection import GridSearchCV
from sklearn.svm import SVC
# Setup the pipeline
steps = [('scaler', StandardScaler()),
('SVM', SVC())]
pipeline = Pipeline(steps)
# Specify the hyperparameter space
parameters = {'SVM__C':[1, 10, 100],
'SVM__gamma':[0.1, 0.01]}
# Create train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=21)
# Instantiate the GridSearchCV object: cv
cv = GridSearchCV(pipeline, parameters, cv=3)
# Fit to the training set
cv.fit(X_train, y_train)
# Predict the labels of the test set: y_pred
y_pred = cv.predict(X_test)
# Compute and print metrics
print("Accuracy: {}".format(cv.score(X_test, y_test)))
print(classification_report(y_test, y_pred))
print("Tuned Model Parameters: {}".format(cv.best_params_))
Accuracy: 0.7795918367346939
precision recall f1-score support
False 0.83 0.85 0.84 662
True 0.67 0.63 0.65 318
micro avg 0.78 0.78 0.78 980
macro avg 0.75 0.74 0.74 980
weighted avg 0.78 0.78 0.78 980
Tuned Model Parameters: {'SVM__C': 10, 'SVM__gamma': 0.1}
Exercise
Bringing it all together II: Pipeline for regression
For this final exercise, you will return to the Gapminder dataset. Guess what? Even this dataset has missing values that we dealt with for you in earlier chapters! Now, you have all the tools to take care of them yourself!
Your job is to build a pipeline that imputes the missing data, scales the features, and fits an ElasticNet to the Gapminder data. You will then tune the l1_ratio of your ElasticNet using GridSearchCV.
All the necessary modules have been imported, and the feature and target variable arrays have been pre-loaded as X and y.
Instruction
- Set up a pipeline with the following steps:
'imputation', which uses theImputer()transformer and the'mean'strategy to impute missing data ('NaN') using the mean of the column.'scaler', which scales the features usingStandardScaler().'elasticnet', which instantiates anElasticNet()regressor.
- Specify the hyperparameter space for the l1l1 ratio using the following notation:
'step_name__parameter_name'. Here, thestep_nameiselasticnet, and theparameter_nameisl1_ratio. - Create training and test sets, with 40% of the data used for the test set. Use a random state of
42. - Instantiate
GridSearchCVwith the pipeline and hyperparameter space. Use 3-fold cross-validation (This is the default, so you don’t have to specify it). - Fit the
GridSearchCVobject to the training set. - Compute R2R2 and the best parameters. This has been done for you, so hit ‘Submit Answer’ to see the results!
# modified/added by Jinny
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use('ggplot')
df = pd.read_csv('https://s3.amazonaws.com/assets.datacamp.com/production/course_2433/datasets/gapminder-clean.csv')
y = df['life'].values
X = df.drop('life', axis=1).values
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline
from sklearn.model_selection import train_test_split
from sklearn.linear_model import ElasticNet
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
numpy.ndarray
# Setup the pipeline steps: steps
# modified by Jinny: Imputer -> SimpleImputer
steps = [('imputation', SimpleImputer(missing_values=np.nan, strategy='mean')),
('scaler', StandardScaler()),
('elasticnet', ElasticNet())]
# Create the pipeline: pipeline
pipeline = Pipeline(steps)
# Specify the hyperparameter space
parameters = {'elasticnet__l1_ratio':np.linspace(0,1,30)}
# Create train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=42)
# Create the GridSearchCV object: gm_cv
gm_cv = GridSearchCV(pipeline, parameters, cv=3)
# Fit to the training set
gm_cv.fit(X_train, y_train)
# Compute and print the metrics
r2 = gm_cv.score(X_test, y_test)
print("Tuned ElasticNet Alpha: {}".format(gm_cv.best_params_))
print("Tuned ElasticNet R squared: {}".format(r2))
Tuned ElasticNet Alpha: {'elasticnet__l1_ratio': 1.0}
Tuned ElasticNet R squared: 0.8862016570888217