R语言缺失值处理
缺失值
1. is.na 确实值位置判断
注意: 缺失值被认为是不可比较的,即便是与缺失值自身的比较。这意味着无法使用比较运算
符来检测缺失值是否存在。例如,逻辑测试myvar == NA的结果永远不会为TRUE。作为
替代,你只能使用处理缺失值的函数(如本节中所述的那些)来识别出R数据对象中的缺
失值。
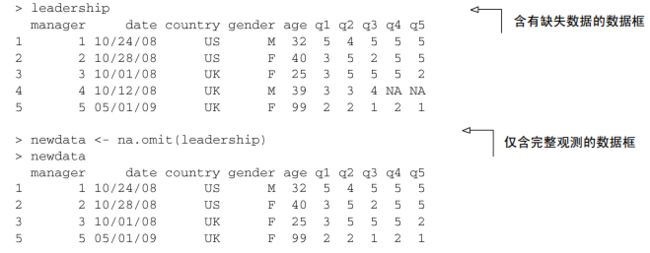
2. na.omit() 删除不完整观测
manyNAs
manyNAs(data, nORp = 0.2)
Arguments
data
A data frame with the data set.
nORp
A number controlling when a row is considered to have too many NA values (defaults to 0.2, i.e. 20% of the columns). If no rows satisfy the constraint indicated by the user, a warning is generated.
按照比例判断缺失.
3. knnImputation K近邻填补
library(DMwR)
knnImputation(data, k = 10, scale = T, meth = "weighAvg", distData = NULL)Arguments
| Arguments | |
|---|---|
| data | A data frame with the data set |
| k | The number of nearest neighbours to use (defaults to 10) |
| scale | Boolean setting if the data should be scale before finding the nearest neighbours (defaults to T) |
| meth | String indicating the method used to calculate the value to fill in each NA. Available values are ‘median’ or ‘weighAvg’ (the default). |
| distData | Optionally you may sepecify here a data frame containing the data set that should be used to find the neighbours. This is usefull when filling in NA values on a test set, where you should use only information from the training set. This defaults to NULL, which means that the neighbours will be searched in data |
Details
This function uses the k-nearest neighbours to fill in the unknown (NA) values in a data set. For each case with any NA value it will search for its k most similar cases and use the values of these cases to fill in the unknowns.
If meth=’median’ the function will use either the median (in case of numeric variables) or the most frequent value (in case of factors), of the neighbours to fill in the NAs. If meth=’weighAvg’ the function will use a weighted average of the values of the neighbours. The weights are given by exp(-dist(k,x) where dist(k,x) is the euclidean distance between the case with NAs (x) and the neighbour k
例子:
#首先读入程序包并对数据进行清理
library(DMwR)
data(algae)
algae <- algae[-manyNAs(algae), ]
clean.algae <- knnImputation(algae[,1:12],k=10) > head(clean.algae)
season size speed mxPH mnO2 Cl NO3 NH4 oPO4 PO4 Chla a1
1 winter small medium 8.00 9.8 60.800 6.238 578.000 105.000 170.000 50.0 0.0
2 spring small medium 8.35 8.0 57.750 1.288 370.000 428.750 558.750 1.3 1.4
3 autumn small medium 8.10 11.4 40.020 5.330 346.667 125.667 187.057 15.6 3.3
4 spring small medium 8.07 4.8 77.364 2.302 98.182 61.182 138.700 1.4 3.1
5 autumn small medium 8.06 9.0 55.350 10.416 233.700 58.222 97.580 10.5 9.2
6 winter small high 8.25 13.1 65.750 9.248 430.000 18.250 56.667 28.4 15.14. centralImputation()中心插值
用非缺失样本的中位数(median)对缺失数据进行插值
data(algae)
cleanAlgae <- centralImputation(algae)
summary(cleanAlgae)5. complete.cases() 寻找完整数据集
x <- airquality[, -1] # x is a regression design matrix
y <- airquality[, 1] # y is the corresponding response
#验证是否complete.cases结果与is.na一样
stopifnot(complete.cases(y) != is.na(y))
#x,y共同的非缺失行的bool结果
ok <- complete.cases(x, y)
#共有几个缺失样本
sum(!ok) # how many are not "ok" ?
#得到非缺失样本
x <- x[ok,]
y <- y[ok]6. na.fail()是否有遗漏值
DF <- data.frame(x = c(1, 2, 3), y = c(0, 10, NA))
na.fail(DF)
Error in na.fail.default(DF) : 对象里有遺漏值