R语言统计入门第六章——回归与相关性
回归与相关性
6.1简单线性回归
library(ISwR)
attach(thuesen)
The following objects are masked from thuesen (pos = 3):
blood.glucose, short.velocity
The following objects are masked from thuesen (pos = 6):blood.glucose, short.velocity
lm(short.velocity~blood.glucose)#线形模型,short.velocity通过blood.glucose来描述
Call:
lm(formula = short.velocity ~ blood.glucose)Coefficients:
(Intercept) blood.glucose
1.09781 0.02196
summary(lm(short.velocity~blood.glucose))#对回归结果进行分析和检验
Call:
lm(formula = short.velocity ~ blood.glucose)Residuals:
Min 1Q Median 3Q Max
-0.40141 -0.14760 -0.02202 0.03001 0.43490
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.09781 0.11748 9.345 6.26e-09 ***
blood.glucose 0.02196 0.01045 2.101 0.0479 *Signif. codes: 0 ‘’ 0.001 '’ 0.01 '’ 0.05 ‘.’ 0.1 ’ ’ 1
Residual standard error: 0.2167 on 21 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.1737, Adjusted R-squared: 0.1343
F-statistic: 4.414 on 1 and 21 DF, p-value: 0.0479
plot(blood.glucose,short.velocity)
abline(lm(short.velocity~blood.glucose))#绘制回归线
缺失值的处理
detach(thuesen)
complete.cases(thuesen)#输出判断缺失值的逻辑向量
[1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[12] TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE
[23] TRUE TRUE
cc<-complete.cases(thuesen)
thuesen
blood.glucose short.velocity
1 15.3 1.76
2 10.8 1.34
3 8.1 1.27
4 19.5 1.47
5 7.2 1.27
6 5.3 1.49
7 9.3 1.31
8 11.1 1.09
9 7.5 1.18
10 12.2 1.22
11 6.7 1.25
12 5.2 1.19
13 19.0 1.95
14 15.1 1.28
15 6.7 1.52
16 8.6 NA
17 4.2 1.12
18 10.3 1.37
19 12.5 1.19
20 16.1 1.05
21 13.3 1.32
22 4.9 1.03
23 8.8 1.12
24 9.5 1.70
thuesen1<-thuesen[cc,]#剔出有缺失值的观测,并赋给thuesen1
thuesen1
blood.glucose short.velocity
1 15.3 1.76
2 10.8 1.34
3 8.1 1.27
4 19.5 1.47
5 7.2 1.27
6 5.3 1.49
7 9.3 1.31
8 11.1 1.09
9 7.5 1.18
10 12.2 1.22
11 6.7 1.25
12 5.2 1.19
13 19.0 1.95
14 15.1 1.28
15 6.7 1.52
17 4.2 1.12
18 10.3 1.37
19 12.5 1.19
20 16.1 1.05
21 13.3 1.32
22 4.9 1.03
23 8.8 1.12
24 9.5 1.70
attach(thuesen1)
The following objects are masked from thuesen (pos = 3):
blood.glucose, short.velocity
The following objects are masked from thuesen (pos = 6):
blood.glucose, short.velocity
lm(short.velocity~blood.glucose)#线形模型,short.velocity通过blood.glucose来描述
Call:
lm(formula = short.velocity ~ blood.glucose)Coefficients:
(Intercept) blood.glucose
1.09781 0.02196
detach(thuesen1)
6.2残差与回归值
attach(thuesen)
The following objects are masked from thuesen (pos = 3):
blood.glucose, short.velocity
The following objects are masked from thuesen (pos = 6):
blood.glucose, short.velocity
lm.velo<-lm(short.velocity~blood.glucose)#将回归结果赋给lm.velo
fitted(lm.velo)#计算回归值
1 2 3 4 5 6 7 8
1.433841 1.335010 1.275711 1.526084 1.255945 1.214216 1.302066 1.341599
9 10 11 12 13 14 15 16
1.262534 1.365758 1.244964 1.212020 1.515103 1.429449 1.244964 NA
17 18 19 20 21 22 23 24
1.190057 1.324029 1.372346 1.451411 1.389916 1.205431 1.291085 1.306459
resid(lm.velo)#输出残差
1 2 3 4 5
0.326158532 0.004989882 -0.005711308 -0.056084062 0.014054962
6 7 8 9 10
0.275783754 0.007933665 -0.251598875 -0.082533795 -0.145757649
11 12 13 14 15
0.005036223 -0.022019994 0.434897199 -0.149448964 0.275036223
16 17 18 19 20
NA -0.070057471 0.045971143 -0.182346406 -0.401411486
21 22 23 24
-0.069916424 -0.175431237 -0.171085074 0.393541161
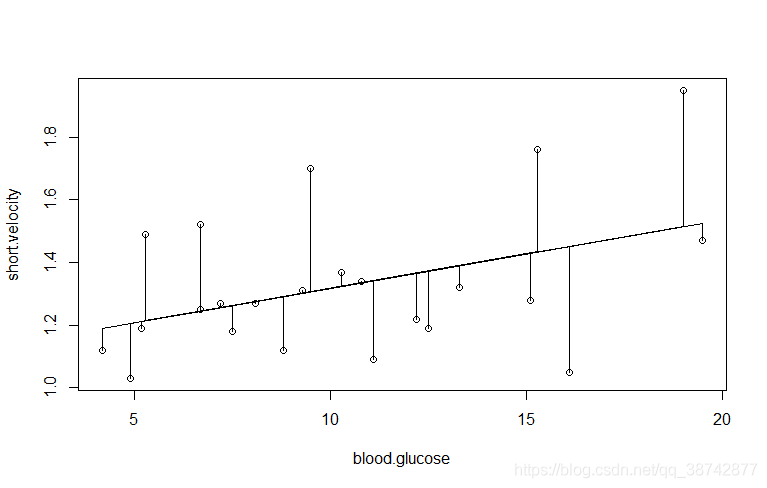
plot(blood.glucose,short.velocity)
lines(blood.glucose,fitted(lm.velo))#绘制回归线,
lines(blood.glucose[!is.na(short.velocity)],fitted(lm.velo))
Error in xy.coords(x, y): ‘x’ and ‘y’ lengths differ
cc<-complete.cases(thuesen)
options(na.action = na.exclude)#缺失值处理选项
clm.velo<-lm(short.velocity~blood.glucose)#不使用上条命令时,将na.action = na.exclude作为参数设置结果一致
fitted(lm.velo)#计算回归值
1 2 3 4 5 6 7 8
1.433841 1.335010 1.275711 1.526084 1.255945 1.214216 1.302066 1.341599
9 10 11 12 13 14 15 16
1.262534 1.365758 1.244964 1.212020 1.515103 1.429449 1.244964 NA
17 18 19 20 21 22 23 24
1.190057 1.324029 1.372346 1.451411 1.389916 1.205431 1.291085 1.306459
segments(blood.glucose,fitted(lm.velo),blood.glucose,short.velocity)#绘制残差线段
plot(fitted(lm.velo),resid(lm.velo))#残差与回归值的散点图
qqnorm(resid(lm.velo))#通过qq图检验残差的正态性
6.3预测与置信带
predict(lm.velo)#不加参数时,predict输出的是回归值
1 2 3 4 5 6 7 8
1.433841 1.335010 1.275711 1.526084 1.255945 1.214216 1.302066 1.341599
9 10 11 12 13 14 15 16
1.262534 1.365758 1.244964 1.212020 1.515103 1.429449 1.244964 NA
17 18 19 20 21 22 23 24
1.190057 1.324029 1.372346 1.451411 1.389916 1.205431 1.291085 1.306459
predict(lm.velo,int="c")#得到自信边界值,fit期望得到的值,lwr下界,upr上界
fit lwr upr
1 1.433841 1.291371 1.576312
2 1.335010 1.240589 1.429431
3 1.275711 1.169536 1.381887
4 1.526084 1.306561 1.745607
5 1.255945 1.139367 1.372523
6 1.214216 1.069315 1.359118
7 1.302066 1.205244 1.398889
8 1.341599 1.246317 1.436881
9 1.262534 1.149694 1.375374
10 1.365758 1.263750 1.467765
11 1.244964 1.121641 1.368287
12 1.212020 1.065457 1.358583
13 1.515103 1.305352 1.724854
14 1.429449 1.290217 1.568681
15 1.244964 1.121641 1.368287
16 NA NA NA
17 1.190057 1.026217 1.353898
18 1.324029 1.230050 1.418008
19 1.372346 1.267629 1.477064
20 1.451411 1.295446 1.607377
21 1.389916 1.276444 1.503389
22 1.205431 1.053805 1.357057
23 1.291085 1.191084 1.391086
24 1.306459 1.210592 1.402326
predict(lm.velo,int="p")#预测边界
Warning in predict.lm(lm.velo, int = “p”): predictions on current data refer to future responses
fit lwr upr
1 1.433841 0.9612137 1.906469
2 1.335010 0.8745815 1.795439
3 1.275711 0.8127292 1.738693
4 1.526084 1.0248161 2.027352
5 1.255945 0.7904672 1.721423
6 1.214216 0.7408499 1.687583
7 1.302066 0.8411393 1.762993
8 1.341599 0.8809929 1.802205
9 1.262534 0.7979780 1.727090
10 1.365758 0.9037136 1.827802
11 1.244964 0.7777510 1.712177
12 1.212020 0.7381424 1.685898
13 1.515103 1.0180367 2.012169
14 1.429449 0.9577873 1.901111
15 1.244964 0.7777510 1.712177
16 NA NA NA
17 1.190057 0.7105546 1.669560
18 1.324029 0.8636906 1.784367
19 1.372346 0.9096964 1.834996
20 1.451411 0.9745421 1.928281
21 1.389916 0.9252067 1.854626
22 1.205431 0.7299634 1.680899
23 1.291085 0.8294798 1.752690
24 1.306459 0.8457315 1.767186
pred.frame<-data.frame(blood.glucose=4:20)#生成blood.glucose的新数据框
pp<-predict(lm.velo,int="p",newdata=pred.frame)#预测pred.frame中的y值,计算预测区间,并赋给pp
pc<-predict(lm.velo,int="c",newdata=pred.frame)#预测pred.frame中的y值,计算自信区间,并赋给pc
plot(blood.glucose,short.velocity,ylim = range(short.velocity,pp,na.rm = T))#绘制散点图,确定图形比例
pred.gluc<-pred.frame$blood.glucose#提取变量数据并赋给新的数据框,
matlines(pred.gluc,pc,lty = c(1,2,2),col="black")
matlines(pred.gluc,pp,lty=c(1,3,3),col="black")
6.4相关性
6.4.1皮尔逊相关系数
cor(blood.glucose,short.velocity)#结果缺失,因为参数存在缺失值
[1] NA
cor(blood.glucose,short.velocity,use="complete.obs")#对blood.glucose,short.velocity进行相关系数计算,use=c为缺失值处理选项
[1] 0.4167546
cor(thuesen,use="complete.obs")#对数据框中所有变量进行相关系数计算生成相关系数矩阵
blood.glucose short.velocity
blood.glucose 1.0000000 0.4167546
short.velocity 0.4167546 1.0000000
cor.test(blood.glucose,short.velocity)#相关性检验
Pearson’s product-moment correlation
data: blood.glucose and short.velocity
t = 2.101, df = 21, p-value = 0.0479
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.005496682 0.707429479
sample estimates:
cor
0.4167546
### 6.4.2斯皮尔曼相关系数
cor.test(blood.glucose,short.velocity,method = "spearman")#斯皮尔曼相关系数检验,method=s为其选项
Warning in cor.test.default(blood.glucose, short.velocity, method =
“spearman”): Cannot compute exact p-value with ties
Spearman’s rank correlation rho
data: blood.glucose and short.velocity
S = 1380.4, p-value = 0.1392
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.318002
6.4.3肯德尔等级相关系数
cor.test(blood.glucose,short.velocity,method = "kendall")#与上同
Warning in cor.test.default(blood.glucose, short.velocity, method =
“kendall”): Cannot compute exact p-value with ties
Kendall’s rank correlation tau
data: blood.glucose and short.velocity
z = 1.5604, p-value = 0.1187
alternative hypothesis: true tau is not equal to 0
sample estimates:
tau
0.2350616