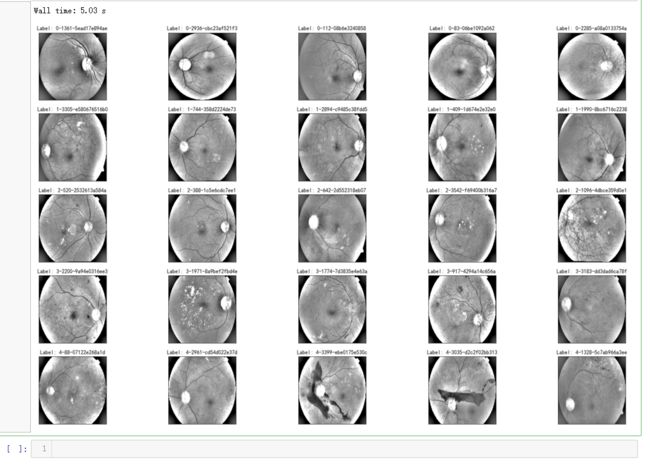
%%time fig = plt.figure(figsize=(25, 16)) for class_id in sorted(train_y.unique()): for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(5, random_state=SEED).iterrows()): ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[]) path="F:\\kaggleDataSet\\diabeticRetinopathy\\resized train 19\\"+str(row['id_code'])+".jpg" image = cv2.imread(path) image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY) # image=cv2.addWeighted ( image, 0 , cv2.GaussianBlur( image , (0 ,0 ) , 10) ,-4 ,128) image = cv2.resize(image, (IMG_SIZE, IMG_SIZE)) plt.imshow(image, cmap='gray') ax.set_title('Label: %d-%d-%s' % (class_id, idx, row['id_code']) )
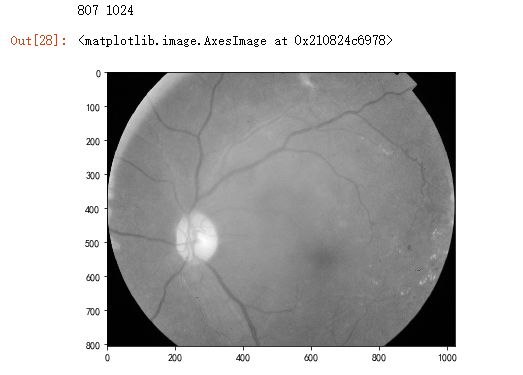
dpi = 80 #inch # path=f"../input/aptos2019-blindness-detection/train_images/5c7ab966a3ee.png" # notice upper part path="F:\\kaggleDataSet\\diabeticRetinopathy\\resized train 19\\cd54d022e37d.jpg" # lower-right, this still looks not so severe, can be class3 image = cv2.imread(path) image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY) height, width = image.shape print(height, width) SCALE=2 figsize = (width / float(dpi))/SCALE, (height / float(dpi))/SCALE fig = plt.figure(figsize=figsize) plt.imshow(image, cmap='gray')
%%time fig = plt.figure(figsize=(25, 16)) for class_id in sorted(train_y.unique()): for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(5, random_state=SEED).iterrows()): ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[]) path="F:\\kaggleDataSet\\diabeticRetinopathy\\resized train 19\\"+str(row['id_code'])+".jpg" image = cv2.imread(path) image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY) # image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB) image = cv2.resize(image, (IMG_SIZE, IMG_SIZE)) image=cv2.addWeighted ( image,4, cv2.GaussianBlur( image , (0,0) , IMG_SIZE/10) ,-4 ,128) # the trick is to add this line plt.imshow(image, cmap='gray') ax.set_title('Label: %d-%d-%s' % (class_id, idx, row['id_code']) )
def crop_image1(img,tol=7): # img is image data # tol is tolerance mask = img>tol return img[np.ix_(mask.any(1),mask.any(0))] def crop_image_from_gray(img,tol=7): if img.ndim ==2: mask = img>tol return img[np.ix_(mask.any(1),mask.any(0))] elif img.ndim==3: gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY) mask = gray_img>tol check_shape = img[:,:,0][np.ix_(mask.any(1),mask.any(0))].shape[0] if (check_shape == 0): # image is too dark so that we crop out everything, return img # return original image else: img1=img[:,:,0][np.ix_(mask.any(1),mask.any(0))] img2=img[:,:,1][np.ix_(mask.any(1),mask.any(0))] img3=img[:,:,2][np.ix_(mask.any(1),mask.any(0))] # print(img1.shape,img2.shape,img3.shape) img = np.stack([img1,img2,img3],axis=-1) # print(img.shape) return img
def load_ben_color(path, sigmaX=10): image = cv2.imread(path) image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB) image = crop_image_from_gray(image) image = cv2.resize(image, (IMG_SIZE, IMG_SIZE)) image=cv2.addWeighted ( image,4, cv2.GaussianBlur( image , (0,0) , sigmaX) ,-4 ,128) return image
%%time NUM_SAMP=7 fig = plt.figure(figsize=(25, 16)) for class_id in sorted(train_y.unique()): for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(NUM_SAMP, random_state=SEED).iterrows()): ax = fig.add_subplot(5, NUM_SAMP, class_id * NUM_SAMP + i + 1, xticks=[], yticks=[]) path="F:\\kaggleDataSet\\diabeticRetinopathy\\resized train 19\\"+str(row['id_code'])+".jpg" image = load_ben_color(path,sigmaX=30) plt.imshow(image) ax.set_title('%d-%d-%s' % (class_id, idx, row['id_code']) )