皮肤识别系统(1)数据下载和数量占比统计
import pandas as pd
import numpy as np
import os
phases = ['train','valid','test']
data_root = os.path.join(os.getcwd(),'data')
data_dir = {
phase:os.path.join(data_root,phase) for phase in phases}
print(data_root)
print(data_dir)
/home/ss-rong/myJupyterNotebook/data
{'train': '/home/ss-rong/myJupyterNotebook/data/train', 'valid': '/home/ss-rong/myJupyterNotebook/data/valid', 'test': '/home/ss-rong/myJupyterNotebook/data/test'}
如果没有数据集,则改为True
download_images = True
download_more_images = True
mount_gdrive = False
下载数据,速度很慢,得科学上网
#def download(url, destination_folder='.'):
# !wget -nc -q --show-progress $url -P $destination_folder
#if download_images:
# for phase in phases:
# download(f'https://s3-us-west-1.amazonaws.com/udacity-dlnfd/datasets/skin-cancer/test.zip', data_root)
将下载的数据解压,tqdm作用是一个进度条插件;with…as…的作用是读取文件路径(与try…catch类似)
import zipfile
from tqdm import tqdm_notebook as tqdm
if download_images:
for phase in phases:
if not os.path.exists(data_dir[phase]):
with zipfile.ZipFile(os.path.join(data_root,f'{phase}.zip'),'r') as myzip:
for file in tqdm(myzip.namelist(),desc=f'Extracting {phase}.zip'):
myzip.extract(member=file,path=data_root)
查看数据文件夹里的组成部分
import glob
#path.split()作用是分割文件路径;os.path.sep指示分隔符'/'
#glob函数可以查找相应的文件,此处是查找data_root文件夹下所有的有关train的文件
classes = [path.split(os.path.sep)[-1] for path in sorted(glob.glob(os.path.join(data_root, 'train', '*')))] if download_images else ['melanoma', 'nevus', 'seborrheic_keratosis']
classes
['melanoma', 'nevus', 'seborrheic_keratosis']
绘制统计图,统计表格的函数(统计classes和phases各有多少图片,占比多少)
import seaborn as sns
import warnings
warnings.filterwarnings("ignore") #使得代码忽略警告(warnings)提示
def print_images_distribution(plot=False):
#图片的分配,数据结构是 竖排为index(train,test,valid) 横排为columns('melanoma', 'nevus', 'seborrheic_keratosis')
image_repartition=pd.DataFrame(index=[d.split(os.path.sep)[-1] for d in data_dir.values()],columns=classes)
#plot_data的数据结构只有一个横排columns('Class', 'Phase', 'Count')
plot_data = pd.DataFrame(columns=['Class', 'Phase', 'Count'])
#这段代码的作用是统计3种classes分别在train valid test数据集中的图片数量
#可以在'草稿.ipynb'的代码里查看plot_data的数据结构
for phase in [d.split(os.path.sep)[-1] for d in data_dir.values()]:
for disease in classes:
count = len(glob.glob(os.path.join(data_root, phase, disease, '*.jpg')))
image_repartition.loc[phase][disease] = count
plot_data = plot_data.append({
'Class': disease, 'Phase': phase, 'Count': count}, ignore_index=True)
#计算各个phases的总数,和各个classes的总数,还有所占的比例,绘制出表格(数据结构)
image_repartition.loc['TOTAL'] = image_repartition.sum(axis=0)
image_repartition['TOTAL'] = image_repartition.sum(axis=1).astype(int)
image_repartition['Ratio'] = np.round(image_repartition.TOTAL / (image_repartition.TOTAL.sum() - image_repartition.TOTAL[-1]), 2)
#绘制统计图形,柱状图。
if plot:
sns.set_style("whitegrid")
sns.barplot(x='Class', y='Count', hue='Phase', data=plot_data, palette='Blues')
return image_repartition
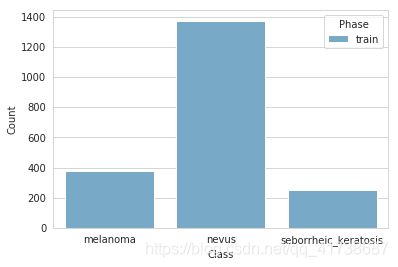
绘制的统计图和统计表如下:
print_images_distribution(True)
| melanoma | nevus | seborrheic_keratosis | TOTAL | Ratio | |
|---|---|---|---|---|---|
| train | 374 | 1372 | 254 | 2000 | 1.0 |
| valid | NaN | NaN | NaN | 0 | 0.0 |
| test | NaN | NaN | NaN | 0 | 0.0 |
| TOTAL | 374 | 1372 | 254 | 2000 | 1.0 |
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-TRSOwylk-1582954353053)(output_12_1.png)]
对比数据的数量不平衡
从上面的数据可以看出nevus(痣)的图片数量很大,但是我们需要的是对melanoma(黑色素瘤)和seborrheic_keratosis(脂溢性角质化病)进行准确的预测; 如果我们不对数据进行更改的话,我们预测痣倒是会更准确,显然这不是我们想要达到的效果
下载更多的数据
我将会从 ISIC Archive.中下载更多的数据,并且确保不会大量重复
#如果我们想通过代码直接下载数据的话,回到前面,把download_more_images赋值为True
if download_more_images:
data_dir['more']=os.path.join(data_root,'more')
data_dir
{'train': '/home/ss-rong/myJupyterNotebook/data/train',
'valid': '/home/ss-rong/myJupyterNotebook/data/valid',
'test': '/home/ss-rong/myJupyterNotebook/data/test',
'more': '/home/ss-rong/myJupyterNotebook/data/more'}
google_drive_download_link = 'https://drive.google.com/uc?export=download'
additional_image_files = {
# additional images downloaded from https://www.isic-archive.com/#!/onlyHeaderTop/gallery
'melanoma-1.zip': '10Wo3RwSC-O4YEAZkU77ZehciDpWCn3Si',
'melanoma-2.zip': '1L9XLBX1sIYX9GiEdfhWizeOuP52Pdgm2',
'melanoma-3.zip': '1jP7YCh5e2g588f0j5ikURsU7oNCLtQCL',
'melanoma-4.zip': '1_IjF2lZZE2yeaZSE9rXqrfrgXd_UdKZi',
'melanoma-5.zip': '1KRF0zRWU-yGSa87p04kQvCHvTiCnZnOO',
'melanoma-6.zip': '1vL18GGPxNCY--R-VLoFRqxeI2ZiSfTJi',
'melanoma-7.zip': '1bFoqLrcUu1YwYq5EQxMrg7WQjRRHNA8U',
'melanoma-8.zip': '1tFW8OkvpWyXpm0ueeCld0qG5k6uk7q3X',
'nevus-1.zip': '1GqBasxjZJNh_C8B4uWaxGLbvDU3HE0e3',
'nevus-2.zip': '1H7PDGPnCapqwmYY2FYaSp1ppqMg9W72a',
'nevus-3.zip': '1snmb9zMdcAjVxW9sRpduMbP9gf3gUqGh',
'nevus-4.zip': '1KFDlx1W8AC97hzTRvVvQJ29EoUiH78pQ',
'seborrheic_keratosis.zip': '1wTgY_AFo0wLF-0g-2mxzkwAv2EXmpGq1',
}
# my own pretrained models
trained_models_files = {
'DenseNet.pt': '1c83tG5-fvTQsRB-N-ADLXwG7_W3mbiwr',
'Inception3_3_0.9051.pt': '1-4-NBGjBzaVLbdKyQYU35JsG7hH9dmpD',
'Inception3_3_0.9089.pt': '1-BE3K5jyDZ21rsFpIG_22GfrCvriLM_G',
'Inception3_1_Mixed_5c.branch3x3dbl_1.bn_0.9126.pt': '1-nDIjuT0FY4UubhM35XCnfAoCVfJD1JI',
'NASNetALarge.pt': '144_os2xtFyN9RDH-1cYiXfRd1p5hA-Gn',
'NASNetALarge_4_0.9106.pt': '1JJLyVraQc8ojllTdU4LyyVo5mkpFtq-3',
#'Xception_1_block4_0.9076.pt': '1WYvzipY-pR5Db6b2bkI8BDfv9HLc1PNt',
'Xception_1_block4_0.9120.pt': '1308ZVjdOsMe_voKRQ5wdejMapa01g1cK',
'Xception_2_block3.rep.4.conv1_0.9185.pt': '1-mjjS7Hgm4knFp5W7XCY3t9DCwCHSAm1',
'SENet_3_layer2.1_0.8988.pt': '1--Q3L7DkuHL3Qr8A6gJZIcP1FiOiMQm_',
'InceptionV4_2_features.6.branch1.0_0.9117.pt': '1-0EXqfAEqO_sqeWW3X7Po6-yRbbVkVyu',
#'InceptionV4_3_features.7_0.9057.pt': '',
#'InceptionV4_6_features.6.branch2_0.8986.pt': '',
'InceptionV4_4_features.6.branch2.1.bn_0.9152.pt': '1-5caaGcjEZEm-FVfGvL-J1imdRR52OOy',
'InceptionResNetV2_2_repeat.0.branch2_0.8966.pt': '1lMsnmDJFoT-xTeUM6UC3axTSaL5f9F0K',
#'PNASNet5Large_1_cell_1.comb_iter_4_left_0.9058.pt': '10IM9Pp5a1T8jdxjPKnzqKMYZKi6kbtOL',
#'PNASNet5Large_0_cell_1_0.9070.pt': '',
'PNASNet5Large_3_cell_0.comb_iter_4_left.separable_2_0.9254.pt': '17OhGCpKNloXs8Oo6UJOT6g3HGKWE421-',
}