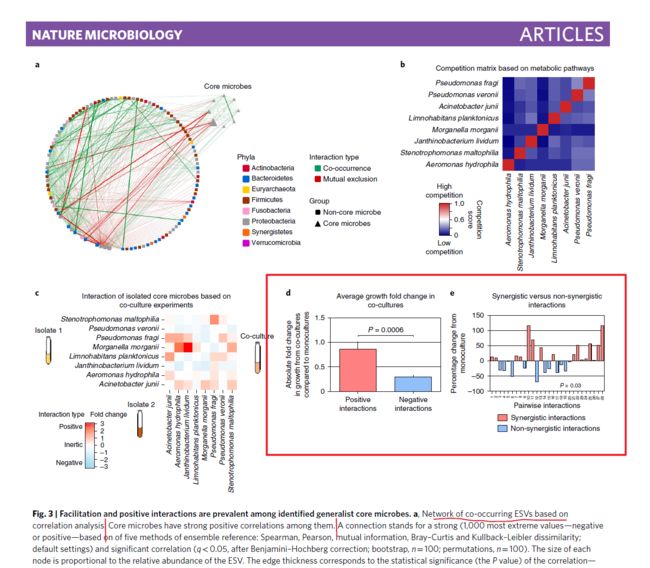
今天要模仿的图片来自于论文 Core gut microbial communities are maintained by beneficial interactions and strain variability in fish。期刊是 Nature microbiology
image.png
今天重复的图片是Figure3中的柱形图
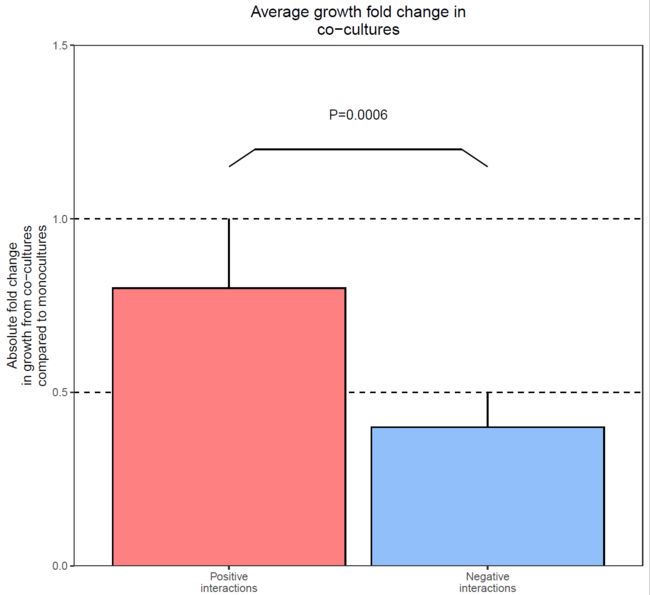
首先是第一个小图:柱形图,误差棒,显著性P值
第一步是模拟数据
数据是三列:第一列用来做X,第二列做Y,第三列做误差条
读入数据
df1<-read.csv("Figure3_d.csv",header=T)
ggplot2基本的柱形图,利用分组信息填充颜色
library(ggplot2)
ggplot(df1,aes(x=group,y=value))+
geom_col(aes(fill=group),color="black")
接下来是简单地美化,包括调整配色,扩大y轴范围,去掉灰色背景,柱子贴底,去掉图例,更改x和y轴的标题,添加总标题,添加辅助线,添加误差线等等。
下面的代码就不详细介绍了,争取录制一期视频来介绍
ggplot(df1,aes(x=group,y=value))+
geom_hline(yintercept = 0.5,lty="dashed")+
geom_hline(yintercept = 1,lty="dashed")+
geom_col(aes(fill=group),color="black")+
theme_bw()+
theme(panel.background = element_blank(),
panel.grid = element_blank(),
plot.title = element_text(hjust=0.5),
legend.position = "none")+
scale_y_continuous(expand = c(0,0),
limits = c(0,1.5))+
scale_x_discrete(labels=c("Positive\ninteractions","Negative\ninteractions"))+
annotate("segment",x=1,y=0.8,xend=1,yend=1)+
annotate("segment",x=2,y=0.4,xend=2,yend=0.5)+
labs(x=NULL,
y="Absolute fold change\nin growth from co-cultures\ncompared to monocultures",
title = "Average growth fold change in\nco-cultures")+
annotate("segment",x=1.1,y=1.2,xend=1.9,yend=1.2)+
annotate("segment",x=1,y=1.15,xend=1.1,yend=1.2)+
annotate("segment",x=1.9,y=1.2,xend=2,yend=1.15)+
annotate("text",x=1.5,y=1.3,label="P=0.0006")+
scale_fill_manual(values = c("#ff8080","#90bff9"))
接下来是第二个小图:有正值和负值的柱形图
第一步还是构造数据
x<-1:28
y<-sample(-100:150,28,replace = F)
df2<-data.frame(x,y)
df2$group<-ifelse(df2$y>0,"A","B")
基本的柱形图
df2$x<-factor(df2$x)
ggplot(df2,aes(x,y))+
geom_col(aes(fill=group),color="black")
接下来是美化:包括去掉背景,更改配色,调整x轴标签的角度等等
df2$group<-factor(df2$group,
labels = c("Synergistic interactions",
"Non-synergistic interactions"))
ggplot(df2,aes(x,y))+
geom_hline(yintercept = -50,lty="dashed")+
geom_hline(yintercept = 50,lty="dashed")+
geom_hline(yintercept = 100,lty="dashed")+
geom_col(aes(fill=group),color="black")+
theme_bw()+
theme(panel.background = element_blank(),
panel.grid = element_blank(),
axis.text.x = element_text(angle = 90,hjust=0.5,
vjust = 0.5),
plot.title = element_text(hjust = 0.5),
legend.position = "bottom",
legend.title = element_blank())+
scale_y_continuous(expand=c(0,0),
limits=c(-100,150),
breaks = c(-100,-50,0,50,100,150))+
labs(x="Pairwise interactions",
y="Percentage change from\nmonoculture",
title = "Synergistic versus non-synergistic\ninteractions")+
scale_fill_manual(values = c("#ff8080","#90bff9"))
最后是拼图
library(cowplot)
pdf("Rplot11.pdf",width = 8,,height = 4)
plot_grid(p1,p2,ncol = 2,nrow=1,labels = c("d","e"))
dev.off()
欢迎大家关注我的公众号
小明的数据分析笔记本