编译WRF及第一次运行记录
Compile and Launch WRF:4.2
-
- O NOTE
- Ⅰ My PC's environments
- Ⅱ Preparation
-
- Ⅱ.1 Install some requirements
- Ⅱ.2 Verify that the Fortran compiler is built properly and that it's compatitble with the C compiler
- Ⅲ Build libraries
-
- Ⅲ.1 Download compressed files
- Ⅲ.2 Set environment variables
- Ⅲ.3 Compile libraries
- Ⅳ Test compatibility of libraries
-
- Ⅳ.1 Download test files
- Ⅳ.2 Fortran + C + NetCDF
- Ⅳ.3 Fortran + C + NetCDF + MPI
- Ⅴ Compile WRF and WPS
- Ⅵ Configure static geography data
- Ⅶ Download real-time data
- Ⅷ Run WPS and WRF
-
- Ⅷ.1 Run WPS
- Ⅷ.2 Create soft links with the input GFS data files
- Ⅷ.3 Run WRF
- Ⅸ References
O NOTE
Before steps below, please ensure that there are at least 40G spare disk space, because the image will be very large after all steps:

If there is a little disk space left, try to change the location of the image.
method reference:【Docker】win10上修改docker的镜像文件存储位置(九)- 通过WSL2修改
Ⅰ My PC’s environments
Windows-home
win-Docker
Docker-CentOS:latest
Ⅱ Preparation
Ⅱ.1 Install some requirements
yum install -y gcc gcc-gfortran gcc-c++ which perl csh git wget make tar m4 libXrender libXext fontconfig
I will use gcc to compile WRF and WPS in the following steps.
check it’s version
gcc -version # ensure version >= 4.6
create directories
cd /opt
mkdir Build_WRF Test
Ⅱ.2 Verify that the Fortran compiler is built properly and that it’s compatitble with the C compiler
download test files
cd Test
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/Fortran_C_tests.tar
tar -xf Fortran_C_test.tar
Test 1
gfortran TEST_1_fortran_only_fixed.f
./a.out
output should be:
SUCCESS test 1 fortran only fixed format
Teat 2
gfortran TEST_2_fortran_only_free.f90
./a.out
output should be:
Assume Fortran 2003: has FLUSH, ALLOCATABLE, derived type, and ISO C Binding
SUCCESS test 2 fortran only free format
Test 3
gcc TEST_3_c_only.c
./a.out
output should be:
SUCCESS test 3 c only
Test 4
gcc -c -m64 TEST_4_fortran+c_c.c
gfortran -c -m64 TEST_4_fortran+c_f.f90
gfortran -m64 TEST_4_fortran+c_f.o TEST_4_fortran+c_c.o
./a.out
output should be:
C function called by Fortran
Values are xx = 2.00 and ii = 1
SUCCESS test 4 fortran calling c
Ⅲ Build libraries
Ⅲ.1 Download compressed files
cd Build_WRF
mkdir LIBRARIES
cd LIBRARIES
mkdir tar untar
cd tar
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/mpich-3.0.4.tar.gz
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/netcdf-4.1.3.tar.gz
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/jasper-1.900.1.tar.gz
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/libpng-1.2.50.tar.gz
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/zlib-1.2.7.tar.gz
# if it reports errors when commpiling WRF
wget https://ftp.gnu.org/gnu/time/time-1.9.tar.gz
Ⅲ.2 Set environment variables
csh # change to Cshell
setenv DIR /opt/Build_WRF/LIBRARIES
setenv CC gcc
setenv CXX g++
setenv FC gfortran
setenv FCFLAGS -m64
setenv F77 gfortran
setenv FFLAGS -m64
##Netcdf
setenv PATH $DIR/netcdf/bin:$PATH
setenv NETCDF $DIR/netcdf
##Mpich
setenv PATH $DIR/mpich/bin:$PATH
##Grib2
setenv GRIB2 $DIR/grib2
setenv JASPERLIB $GRIB2/lib
setenv JASPERINC $GRIB2/include
setenv LDFLAGS -L$GRIB2/lib
setenv CPPFLAGS -I$GRIB2/include
setenv LD_LIBRARY_PATH $GRIB2/lib:$LD_LIBRARY_PATH
setenv LD_LIBRARY_PATH $NETCDF/lib:$LD_LIBRARY_PATH
Ⅲ.3 Compile libraries
# NetCDF
cd /opt/Build_WRF/LIBRARIES/tar
tar -xzf netcdf-4.1.3.tar.gz -C /opt/Build_WRF/LIBRARY/untar
cd ../untar/netcdf-4.1.3
./configure --prefix=$DIR/netcdf --disable-dap --disable-netcdf-4 --disable-shared
make
make install
setenv PATH $DIR/netcdf/bin:$PATH
setenv NETCDF $DIR/netcdf
# mpich
tar -zxf mpich-3.0.4.tar.gz -C /opt/Build_WRF/LIBRARIES/untar
cd ../untar/mpich-3.0.4
./configure --prefix=$DIR/mpich
make
make install
setenv PATH $DIR/mpich/bin:$PATH
# zlib
tar -xzf zlib-1.2.7.tar.gz -C /opt/Build_WRF/LIBRARIES/untar
cd ../untar/zlib-1.2.7
./configure --prefix=$DIR/grib2
make
make install
# libpng
tar -xzf libpng-1.2.50.tar.gz -C /opt/Build_WRF/LIBRARIES/untar
cd ../untar/libpng-1.2.50
./configure --prefix=$DIR/grib2
make
make install
# JasPer
tar -xzf jasper-1.900.1.tar.gz -C /opt/Build_WRF/LIBRARIES/untar
cd ../untar/jasper-1.900.1
./configure --prefix=$DIR/grib2
make
make install
# if it reports errors when commpiling WRF
# time
tar -xzf time-1.9.tar.gz -C /opt/Build_WRF/LIBRARIES/untar
cd ../untar/time-1.9
./configure -–prefix=/usr/
make
make install
ls /usr/bin/time # check whether it was installed successfully
Ⅳ Test compatibility of libraries
Ⅳ.1 Download test files
cd /opt/TESTS
wget https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/Fortran_C_NETCDF_MPI_tests.tar
tar -xf Fortran_C_NETCDF_MPI_tests.tar
Ⅳ.2 Fortran + C + NetCDF
cp ${NETCDF}/include/netcdf.inc .
gfortran -c 01_fortran+c+netcdf_f.f
gcc -c 01_fortran+c+netcdf_c.c
gfortran 01_fortran+c+netcdf_f.o 01_fortran+c+netcdf_c.o -L${NETCDF}/lib -lnetcdff -lnetcdf
./a.out
output should be:
C function called by Fortran
Values are xx = 2.00 and ii = 1
SUCCESS test 1 fortran + c + netcdf
Ⅳ.3 Fortran + C + NetCDF + MPI
cp ${NETCDF}/include/netcdf.inc .
mpif90 -c 02_fortran+c+netcdf+mpi_f.f
mpicc -c 02_fortran+c+netcdf+mpi_c.c
mpif90 02_fortran+c+netcdf+mpi_f.o 02_fortran+c+netcdf+mpi_c.o -L${NETCDF}/lib -lnetcdff -lnetcdf
mpirun ./a.out
output should be:
C function called by Fortran
Values are xx = 2.00 and ii = 1
status = 2
SUCCESS test 2 fortran + c + netcdf + mpi
Ⅴ Compile WRF and WPS
I followed teacher’s advices on choosing option(s) for compilation.
First, compile WRF:
git clone https://github.com/wrf-model/WRF
cd WRF
./configure # choose 34 then 1
./compile em_real >& log.compile
ls -lh main/*.exe
if the run is successful, it will show below:

If WRF has been compiled successfully, then to compile WPS:
cd WPS
./clean
setenv JASPERLIB $DIR/grib2/lib
setenv JASPERINC $DIR/grib2/include
./configure # choose 1
./compile >& log.compile
ls -lh *.exe
If the run is successful, it will show below:
Ⅵ Configure static geography data
cd /opt/Build_WRF/
wget https://www2.mmm.ucar.edu/wrf/src/wps_files/geog_high_res_mandatory.tar.gz
tar -xzf geog_high_res_mandatory.tar.gz # ==> WPS_GEO/
cd /opt/Build_WRF/WPS
vi namelist.wps
change geog_data_path = ´opt/Build_WRF/WPS_GEOG´
Ⅶ Download real-time data
Let’s run an example below.
Note: NOMADS server only displays recently data.
so you should download data depending on yourself conditions.
cd /opt/Build_WRF
mkdir DATA/GFS
cd DATA/GFS
cat > filelist
https://nomads.ncep.noaa.gov/pub/data/nccf/com/gfs/prod/gfs.20210327/00/atmos/gfs.t00z.pgrb2.0p50.f000
https://nomads.ncep.noaa.gov/pub/data/nccf/com/gfs/prod/gfs.20210327/00/atmos/gfs.t00z.pgrb2.0p50.f003
# then press ctrl+D
wget – i filelist
Anyway, you had better rename the GFS data files, according to the date to discriminate what they means. For example, use mvor shell scripts to rename gfs.t00z.pgrb2.0p50.f000 to gfs_20210327_00.grib2 at this directory.
mv gfs.t00z.pgrb2.0p50.f000 gfs_20210327_00.grib2
Ⅷ Run WPS and WRF
Ⅷ.1 Run WPS
Before running geogrid, make sure the geog_data_path in namelist.wps is correct.
cd /opt/Build_WRF/WPS
./geogrid.exe >& log.geogrid
cat log.geogrid
if the run is successful, it will show below:

Ⅷ.2 Create soft links with the input GFS data files
make soft links of the GFS data files to current directory
./link_grib.csh /opt/Build_WRF/DATA/GFS/* # [path_where_you_placed_GFS_files not directory!!!]
make a soft link of the Vtable(Variables Table) of GFS to current directory
ln -sf ungrib/Variable_Tables/Vtable.GFS Vtable
ls -l
output:
![]()
ungrib the grib2 files
./ungrib.exe
if the run is successful, it will show below:
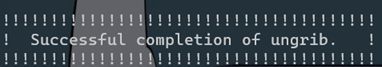
generate FIFLE*:
![]()
if it reports an error that
ERROR: Grib2 file or date problem, stopping in edition_num.
First, check whether the suffix of the files is.grib, because WPS can only read the file with suffix of.grib, instead of.nc.
Then, check whether the soft links link the specific data files, instead of directory.
Finally, check the size of the files, They are certainly not too small.
./metgrid.exe >& log.metgrid
if the run is successful, it will show below:
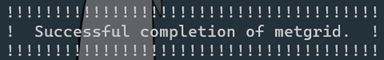
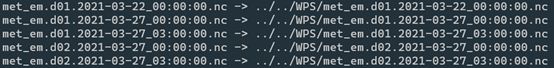
and generate files met_em*:
if it reports an error that
ERROR: Error in ext_pkg_write_field
please updateVtable.GFS, link below
https://www2.mmm.ucar.edu/wrf/src/Vtable.GFS_new
Ⅷ.3 Run WRF
cd ../WRF/run
or
cd ../WRF/test/em_real
you need to make all necessary changes to reflect your particular case to the namelist.input file (t’s important). Once that is complete, you need to copy or link your met_em* files into the working directory:
ln -sf or cp
ln -sf ../../WPS/met_em* . (from the run/ directory) or
ln -sf ../../../WPS/met_em* . (from the test/em_real directory)
generate met_em*:
mpirun -np 1 ./real.exe
generate wrfbdy_01, wrfinput*:

chek rsl to make sure the run is successful:
tail rsl.error.0000
if the run is successful, it will show below:
![]()
run wrf
mpirun -np 8 ./wrf.exe
tail rsl.error.0000
if the run is successful, it will show below:![]()
OK! you can enjoy your WRF data now!
The last, as for the NCL, I command use miniconda to install it.
cd ~
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
# then
./Miniconda3-latest-Linux-x86_64.sh
if you install miniconda successfully, then create an env:
conda create -n ncl_stable -c conda-forge ncl
after installing, type:
conda activate ncl_stable
ncl
to use ncl
PS: I have pushed some images containing WRF to Docker Hub
you can have a try directly by pulling them, which equals skipping compilation or other steps.
By the way, the WRF main directory is /opt/Build_WRF
Ⅸ References
[1] WRF Online Tutorial
[2] 【Docker】win10上修改docker的镜像文件存储位置(九)- 通过WSL2修改
[3] Windows 10 将 Docker Desktop for Windows(WSL 2 方式)文件存储移出系统盘放置到其它目录
[4] 在WSL环境下使用IPython进行Python版NCL的编程