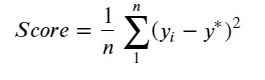task09:集成学习案例——蒸汽量预测
准备工作
背景介绍
使用的是经脱敏后的锅炉传感器采集的数据(采集频率是分钟级别)。根据锅炉的工况,预测产生的蒸汽量。
数据信息
数据分成训练数据(train.txt)和测试数据(test.txt),其中字段”V0”-“V37”,这38个字段是作为特征变量,”target”作为目标变量。我们需要利用训练数据训练出模型,预测测试数据的目标变量。
评价指标
代码实战
导入包
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import seaborn as sns
# 模型
import pandas as pd
import numpy as np
from scipy import stats
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV, RepeatedKFold, cross_val_score,cross_val_predict,KFold
from sklearn.metrics import make_scorer,mean_squared_error
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet
from sklearn.svm import LinearSVR, SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor,AdaBoostRegressor
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures,MinMaxScaler,StandardScaler
数据读取
data_train = pd.read_csv('train.txt',sep = '\t')
data_test = pd.read_csv('test.txt',sep = '\t')
#合并训练数据和测试数据
data_train["oringin"]="train"
data_test["oringin"]="test"
data_all=pd.concat([data_train,data_test],axis=0,ignore_index=True)
数据预处理
探索数据分布,传感器是连续变量,所以使用 kdeplot(核密度估计图) 进行数据的初步分析,即EDA。
notes:核密度估计(kernel density estimation):是在概率论中用来估计未知的密度函数,属于非参数检验方法之一。通过核密度估计图可以比较直观的看出数据样本本身的分布特征。
#输出v1-v37的核密度估计图
for column in data_all.columns[0:-2]:
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "train")], color="Red", shade = True)
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "test")], ax =g, color="Blue", shade= True)
g.set_xlabel(column)
g.set_ylabel("Frequency")
g = g.legend(["train","test"])
plt.show()
特征"V5",“V9”,“V11”,“V17”,“V22”,"V28"中训练集数据分布和测试集数据分布不均,删除这些特征数据
for column in ["V5","V9","V11","V17","V22","V28"]:
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "train")], color="Red", shade = True)
g = sns.kdeplot(data_all[column][(data_all["oringin"] == "test")], ax =g, color="Blue", shade= True)
g.set_xlabel(column)
g.set_ylabel("Frequency")
g = g.legend(["train","test"])
plt.show()
data_all.drop(["V5","V9","V11","V17","V22","V28"],axis=1,inplace=True)
特征绘图,观察特征的变化分布情况。
data_train1=data_all[data_all["oringin"]=="train"].drop("oringin",axis=1)
fcols = 2
frows = len(data_train.columns)
plt.figure(figsize=(5*fcols,4*frows))
i=0
for col in data_train1.columns:
i+=1
ax=plt.subplot(frows,fcols,i)
sns.regplot(x=col, y='target', data=data_train, ax=ax,
scatter_kws={
'marker':'.','s':3,'alpha':0.3},
line_kws={
'color':'k'});
plt.xlabel(col)
plt.ylabel('target')
i+=1
ax=plt.subplot(frows,fcols,i)
sns.distplot(data_train[col].dropna() , fit=stats.norm)
plt.xlabel(col)
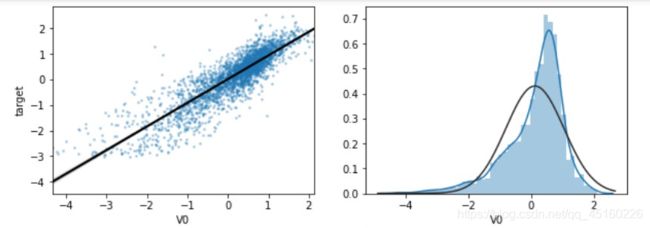
data_train1=data_all[data_all["oringin"]=="train"].drop("oringin",axis=1)
plt.figure(figsize=(20, 16)) # 指定绘图对象宽度和高度
colnm = data_train1.columns.tolist() # 列表头
mcorr = data_train1[colnm].corr(method="spearman") # 相关系数矩阵,即给出了任意两个变量之间的相关系数
mask = np.zeros_like(mcorr, dtype=np.bool) # 构造与mcorr同维数矩阵 为bool型
mask[np.triu_indices_from(mask)] = True # 角分线右侧为True
cmap = sns.diverging_palette(220, 10, as_cmap=True) # 返回matplotlib colormap对象,调色板
g = sns.heatmap(mcorr, mask=mask, cmap=cmap, square=True, annot=True, fmt='0.2f') # 热力图(看两两相似度)
plt.show()
DataFrame.corr(method=‘pearson’, min_periods=1)
计算数值列的两两相关,不包括NA或者null值,注意,也不包括非数值特征列,例如分类特征。相关系数的变化范围是从-1到1,越接近1,表示有越强的正相关;系数越接近-1表示有越强的负相关;系数越接近0表示两属性之间没有线性相关性,注意,没有线性相关性不代表没有其他非线性相关性。函数返回的是一个相关性矩阵,正对角线的数值均为1,且为对称矩阵。
method : {‘pearson’, ‘kendall’, ‘spearman’}
pearson : standard correlation coefficient
kendall : Kendall Tau correlation coefficient
spearman : Spearman rank correlation
pandas.plotting.scatter_matrix(frame, alpha=0.5, figsize=None, ax=None, grid=False, diagonal=‘hist’, marker=’.’, density_kwds=None, hist_kwds=None, range_padding=0.05, **kwargs)
通过corr()可以得到线性相关性的数值关系,不够形象,通常可以在使用corr之后再使用scatter_matrix绘制出图像,通过散点图更加直观的看见属性之间的联系
threshold = 0.1 #设置阈值
corr_matrix = data_train1.corr().abs()
drop_col=corr_matrix[corr_matrix["target"]<threshold].index
data_all.drop(drop_col,axis=1,inplace=True)
进行归一化操作,把数据映射到0~1范围之内处理,更加便捷快速。
cols_numeric=list(data_all.columns)
cols_numeric.remove("oringin")
def scale_minmax(col):
return (col-col.min())/(col.max()-col.min())
scale_cols = [col for col in cols_numeric if col!='target']
data_all[scale_cols] = data_all[scale_cols].apply(scale_minmax,axis=0)
data_all[scale_cols].describe()
特征工程
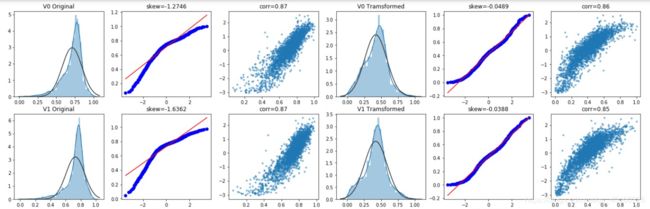
绘图显示Box-Cox变换对数据分布影响,Box-Cox用于连续的响应变量不满足正态分布的情况。在进行Box-Cox变换之后,可以一定程度上减小不可观测的误差和预测变量的相关性。
quantitle-quantile(q-q)图https://blog.csdn.net/u012193416/article/details/83210790
fcols = 6
frows = len(cols_numeric)-1
plt.figure(figsize=(4*fcols,4*frows))
i=0
for var in cols_numeric:
if var!='target':
dat = data_all[[var, 'target']].dropna()
i+=1
plt.subplot(frows,fcols,i)
sns.distplot(dat[var] , fit=stats.norm);
plt.title(var+' Original')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(dat[var], plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(dat[var])))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(dat[var], dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(dat[var], dat['target'])[0][1]))
i+=1
plt.subplot(frows,fcols,i)
trans_var, lambda_var = stats.boxcox(dat[var].dropna()+1)
trans_var = scale_minmax(trans_var)
sns.distplot(trans_var , fit=stats.norm);
plt.title(var+' Tramsformed')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(trans_var, plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(trans_var)))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(trans_var, dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(trans_var,dat['target'])[0][1]))
# 进行Box-Cox变换
cols_transform=data_all.columns[0:-2]
for col in cols_transform:
# transform column
data_all.loc[:,col], _ = stats.boxcox(data_all.loc[:,col]+1)
print(data_all.target.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_all.target.dropna() , fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_all.target.dropna(), plot=plt)
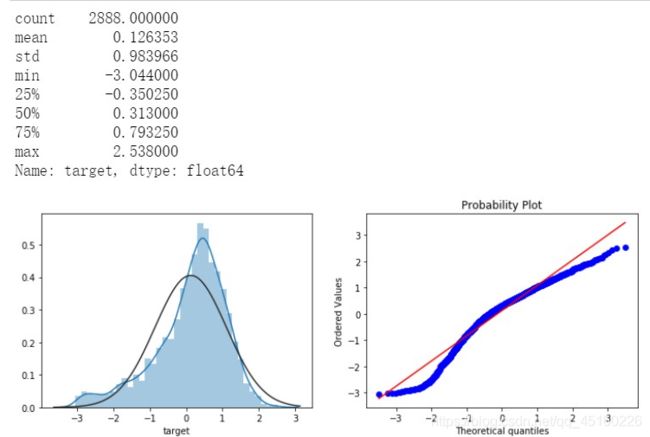
经过Box-Cox变换数据分布,更加正态化,所以进行Box-Cox变换很有必要。Box-Cox变换是Box和Cox在1964年提出的一种广义幂变换方法,是统计建模中常用的一种数据变换,用于连续的响应变量不满足正态分布的情况。
模型构建以及集成学习
# function to get training samples
def get_training_data():
# extract training samples
from sklearn.model_selection import train_test_split
df_train = data_all[data_all["oringin"]=="train"]
df_train["label"]=data_train.target1
# split SalePrice and features
y = df_train.target
X = df_train.drop(["oringin","target","label"],axis=1)
X_train,X_valid,y_train,y_valid=train_test_split(X,y,test_size=0.3,random_state=100)
return X_train,X_valid,y_train,y_valid
# extract test data (without SalePrice)
def get_test_data():
df_test = data_all[data_all["oringin"]=="test"].reset_index(drop=True)
return df_test.drop(["oringin","target"],axis=1)
rmse(均方根误差)、mse(均方误差)的评价函数
from sklearn.metrics import make_scorer
# metric for evaluation
def rmse(y_true, y_pred):
diff = y_pred - y_true
sum_sq = sum(diff**2)
n = len(y_pred)
return np.sqrt(sum_sq/n)
def mse(y_ture,y_pred):
return mean_squared_error(y_ture,y_pred)
# scorer to be used in sklearn model fitting
rmse_scorer = make_scorer(rmse, greater_is_better=False)
#输入的score_func为记分函数时,该值为True(默认值);输入函数为损失函数时,该值为False
mse_scorer = make_scorer(mse, greater_is_better=False)
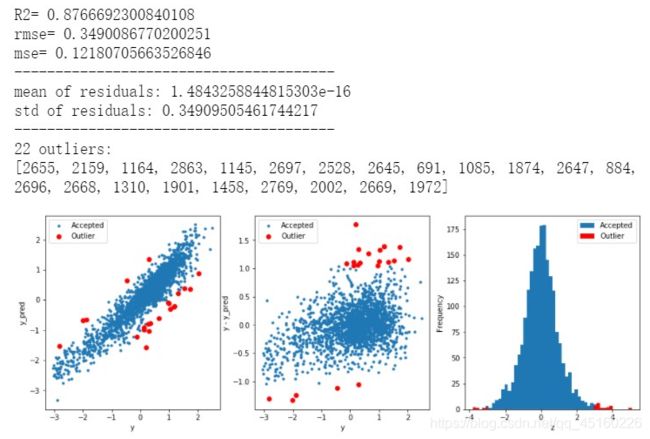
寻找离群值,并删除
# function to detect outliers based on the predictions of a model
def find_outliers(model, X, y, sigma=3):
# predict y values using model
model.fit(X,y)
y_pred = pd.Series(model.predict(X), index=y.index)
# calculate residuals between the model prediction and true y values
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
# calculate z statistic, define outliers to be where |z|>sigma
z = (resid - mean_resid)/std_resid
outliers = z[abs(z)>sigma].index
# print and plot the results
print('R2=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print("mse=",mean_squared_error(y,y_pred))
print('---------------------------------------')
print('mean of residuals:',mean_resid)
print('std of residuals:',std_resid)
print('---------------------------------------')
print(len(outliers),'outliers:')
print(outliers.tolist())
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')
plt.plot(y.loc[outliers],y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y_pred');
ax_132=plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.plot(y.loc[outliers],y.loc[outliers]-y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y - y_pred');
ax_133=plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)
z.loc[outliers].plot.hist(color='r',bins=50,ax=ax_133)
plt.legend(['Accepted','Outlier'])
plt.xlabel('z')
return outliers
# get training data
X_train, X_valid,y_train,y_valid = get_training_data()
test=get_test_data()
# find and remove outliers using a Ridge model
outliers = find_outliers(Ridge(), X_train, y_train)
X_outliers=X_train.loc[outliers]
y_outliers=y_train.loc[outliers]
X_t=X_train.drop(outliers)
y_t=y_train.drop(outliers)
def get_trainning_data_omitoutliers():
#获取训练数据省略异常值
y=y_t.copy()
X=X_t.copy()
return X,y
def train_model(model, param_grid=[], X=[], y=[],
splits=5, repeats=5):
# 获取数据
if len(y)==0:
X,y = get_trainning_data_omitoutliers()
# 交叉验证
rkfold = RepeatedKFold(n_splits=splits, n_repeats=repeats)
# 网格搜索最佳参数
if len(param_grid)>0:
gsearch = GridSearchCV(model, param_grid, cv=rkfold,
scoring="neg_mean_squared_error",
verbose=1, return_train_score=True)
# 训练
gsearch.fit(X,y)
# 最好的模型
model = gsearch.best_estimator_
best_idx = gsearch.best_index_
# 获取交叉验证评价指标
grid_results = pd.DataFrame(gsearch.cv_results_)
cv_mean = abs(grid_results.loc[best_idx,'mean_test_score'])
cv_std = grid_results.loc[best_idx,'std_test_score']
# 没有网格搜索
else:
grid_results = []
cv_results = cross_val_score(model, X, y, scoring="neg_mean_squared_error", cv=rkfold)
cv_mean = abs(np.mean(cv_results))
cv_std = np.std(cv_results)
# 合并数据
cv_score = pd.Series({
'mean':cv_mean,'std':cv_std})
# 预测
y_pred = model.predict(X)
# 模型性能的统计数据
print('----------------------')
print(model)
print('----------------------')
print('score=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print('mse=',mse(y, y_pred))
print('cross_val: mean=',cv_mean,', std=',cv_std)
# 残差分析与可视化
y_pred = pd.Series(y_pred,index=y.index)
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
z = (resid - mean_resid)/std_resid
n_outliers = sum(abs(z)>3)
outliers = z[abs(z)>3].index
return model, cv_score, grid_results
# 定义训练变量存储数据
opt_models = dict()
score_models = pd.DataFrame(columns=['mean','std'])
splits=5
repeats=5
model = 'Ridge' #可替换,见案例分析一的各种模型
opt_models[model] = Ridge() #可替换,见案例分析一的各种模型
alph_range = np.arange(0.25,6,0.25)
param_grid = {
'alpha': alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),
abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')
# 预测函数
def model_predict(test_data,test_y=[]):
i=0
y_predict_total=np.zeros((test_data.shape[0],))
for model in opt_models.keys():
if model!="LinearSVR" and model!="KNeighbors":
y_predict=opt_models[model].predict(test_data)
y_predict_total+=y_predict
i+=1
if len(test_y)>0:
print("{}_mse:".format(model),mean_squared_error(y_predict,test_y))
y_predict_mean=np.round(y_predict_total/i,6)
if len(test_y)>0:
print("mean_mse:",mean_squared_error(y_predict_mean,test_y))
else:
y_predict_mean=pd.Series(y_predict_mean)
return y_predict_mean
进行模型的预测以及结果的保存
y_ = model_predict(test)
y_.to_csv('predict.txt',header = None,index = False)