Genome-Wide Analysis of Prognostic lncRNAs, miRNAs, and mRNAs Forming a Competing Endogenous RNA Network in Hepatocellular Carcinoma
学习一篇文章中的思路
一些生词
univariate 单变量
multivariate 多因素
onset 开始
hint 提示
warrant根据
competing endogenous RNAs (ceRNAs)
To analyze mature miRNAs rather than pre-miRNAs, we downloaded miRNA expression profiles from UCSC Xena (http://xena.ucsc.edu/).
一些文章中可供借鉴的信息
We removed patients with less than 90 days of OS.
Low-abundance genes were also removed when analyzing the relationships between survival and the gene expression profiles.
Genes included in our model were closely related with tumor burden, histological grade, and stage. Importantly, these findings provided several clues that the PI may be particularly useful for the assessment of treatment response and recurrence surveillance.
Cross-talk is thought to occur between lncRNAs, miRNAs, and mRNAs during all aspects of the HCC process, including metastasis , proliferation and invasion.
文章的杂志虽然已经被挪出sci了,但是思路是可以借鉴还有图图是可以学习复现的。
- 下面这个图不知道如何画,就是三种RNA挑选出保护和风险因子,最后挑选出 As a result, we detected a total of 77 survival associated lncRNAs, 29 survival associated miRNAs, and 1014 survival associated。
- The top 20 most significant survival associated genes are presented in Fig. 3
接下来把 The most significant survival associated mRNAs in HCC (P-value < 0.001) were piped to STRING to generate gene interaction networks, and Cytoscape analysis of the gene networks revealed the important cancer pathways, including hub genes, such as CDK1, CCNB1, and PLK1
These most significant survival associated lncRNAs, miRNAs and mRNAs were further submitted to multivariate COX regression analysis to identify the independent prognostic predictors for HCC.
接下来涉及一个公式:用的是3个lncRNAs,3个miRNA,3个mRNAs。问题是这3个是哪来的,是从上面的PPI data筛选来的。3个lncRNAs,3个miRNA,3个mRNAs在公式中的计算如下图所示,3个mRNAs分别是SOCS2、SLC16A11、KPNA2.
By using the three signatures, we calculated a risk score for each patient individually and ranked them according to increased risk score. HCC patients were separated into low- or high-risk groups according to the median risk score. 就是说根据这个计算出的medianrisk score值将HCC病人分成了高、低风险组。
然后,经常在文章中看到的K-M anlysis展示了生存关系:K-M analysis showed a significant difference in OS between the two groups with all three models 80(Fig. 5).
ROC curves比较了预后模型的效率:ROC curves applied to compare the efficiency of these prognostic models.
AUC valuesof
0.728, 0.719, and 0.724, respectively (Fig. 6). 这个
AUC values是评价这个模型的可靠性嘛?
这个FIG.6涉及四个概念性问题:
ROC curves
AUC values
Prognostic index(PI)
Time-dependent ROC curves analysis 是怎么得出的?
-
As shown in Fig. 7, the scores assigned to each patient provide a good assessment of prognosis.
Fig. 7. Performance of prognostic signatures in distinguishing patients into high- and low-risk groups. Figure 7
(A) The distribution of lncRNA-based risk score.
(B) The distribution of miRNA-based risk score.
(C) The distribution of mRNA-based risk score.The Y axis of panel A-C represent the risk score of each patient. The X axis of panel A-C represent the rank of each patient based on risk score.
(D) lncRNA-based risk score for the distribution of patients’ survival status.
(E) miRNA-based risk score for the distribution of patients’ survival status.
(F) mRNA-based risk score for the distribution of patients’ survival status.The Y axis of panel D-F represent the survival time of each patients. The X axis of panel D-F represent the rank of each patient based on risk score.
The expression pattern of these RNAs in the high- and low-risk groups are also displayed (Fig. 7G-I).
-
可看一下下面的图解释,高低风险组分别有哪些基因是高表达,哪些基因是低表达。然后可以反过来解释就是在这个病人检查出的指标,如果这些基因高表达意味着你是高风险组,如果这些基因高表达因为如果是低风险组,那么即使你这些基因高表达也没关系。或者因为你这些基因的低表达就是意味着是高风险组,所以你的预后并不会好。
(G) TMCC1-AS1, LINC01138 and AC009005.1 are significantly up-regulated in the high-risk group according to lncRNA-based risk score.
(H) Mir-9-5P and mir-326 are significantly up-regulated in the high-risk group, while mir-139-5P is down-regulated in the high-risk group according to miRNA-based risk score.
(I) KPNA2 is significantly up-regulated in the high-risk group, while SOCS2 and SLC16A11 is down- regulated in the high-risk group according to mRNA-based risk score.
然后接下来作者说这个包含不同类型RNA的风险预测器可以揭示HCC的潜在分子机制并且增加预测准确性。
Finally, the multiple types of RNA-based PI for HCC OS were selected, including the above nine RNAs. To determine the value of the PI in predicting HCC prognosis, we performed K-M analysis which revealed that patients in the high-risk group had a shorter OS (hazard ratio [HR] = 4.030, 95% confidence interval [CI]: 2.753-5.898, P-value < 0.001; Fig. 8A
the AUC of the time-dependent ROC curve was 0.776 Fig. 8B
To display the prognostic efficiency of PI more intuitively, we also generated a K-M plot of tumor stage and the corresponding ROC curve. Tumor, node, metastasis (TNM) stage also divided the HCC patients into two groups with a significant difference in prognosis (HR=3.897, 95%CI: 2.399- 6.271, P-value < 0.001; Fig. 8C)
The AUC of the ROC curve based on TNM stage was 0.668 Fig. 8D
这里面Fig 8又涉及了一个概念就是
A图:K-M和PI(prognostic index)的关系
B图:Time-dependent ROC curve 与AUC of PI(prognostic index)的关系
- 计算这九个分子和临床性状的相关性。作者在文中这样描述的过渡
- Therefore, we analyzed the expression of these genes under various clinical parameters and investigated their associations with clinical progress. We specifically analyzed several major clinical parameters, including age (over or under 60 years), gender (male or female), tumor status (with tumor or tumor free), pathologic stage (Stage III, IV or Stage I, II), T stage (Stage III, IV or Stage I, II), N stage (whether lymphatic metastasis had occurred or not), M stage (whether distant metastasis had occurred or not), and histological grade (III, IV or I, II). The nine genes included in our risk score showed significant correlation with HCC tumor burden, histological grade, and stage (Fig. 9, Tables 2-4). These results revealed that these genes can be used for effective risk stratification in HCC
疑问:接下来作者列出了一个表格,展示如下,不过坐标的这几项的人数n是不一样的多的,也就是说作者是单独拿出来某一项来做的嘛,然后上面的热图是组后组合在一起的嘛?
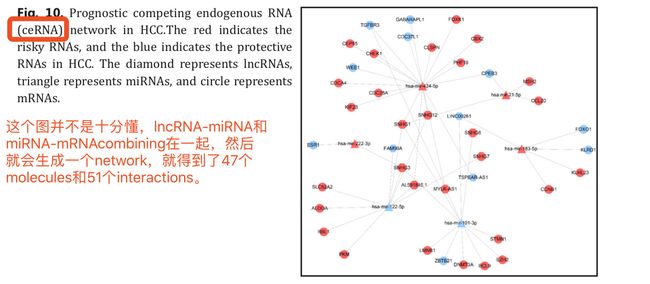
- 接下来Survival-related ceRNA network in HCC ==Combining the lncRNA-miRNA interactions with the miRNA-mRNA interactions==, an integrated lncRNA- miRNA-mRNA network was established, consisting of 47 molecules and 51 interactions Fig. 10
-
Our ceRNA network provide some novel insights into the diagnosis, surveillance, and prognosis of HCC. Considering the regulatory role of lncRNAs, K-M plots generated by Cutoff Finder [21] were used to show their value in predicting prognosis (Fig. 11)
上面这个FIG11用的画的这些生存分析曲线是基于上面的网络图
文章提到一个cutoff finder,老大也写过一个生存分析是一个认人打扮的小姑娘
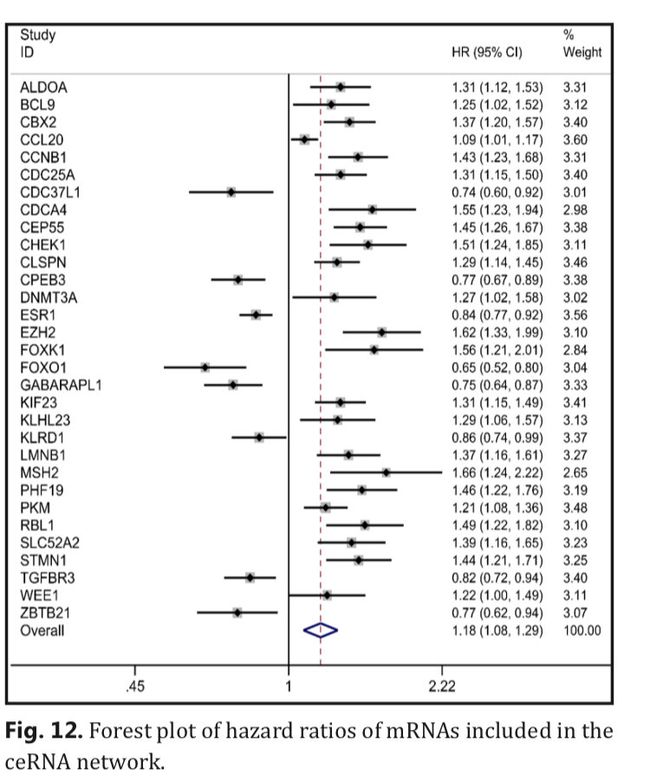
Thus, we performed a comprehensive integrating analysis to assess whether the mRNAs included in the ceRNA network correlated with the prognosis of HCC patients as a whole. It showed that the overexpression of these mRNAs was significantly associated with poor OS (pooled HR=1.18, 95% CI: 1.08– 1.29, P-value < 0.001, random effect; Fig. 12
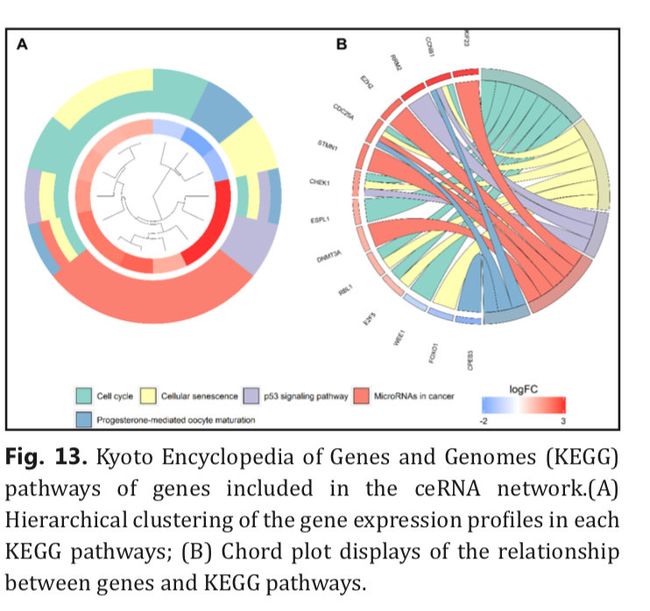
最后作者做了KEGG和GO分析,但是图是长下面这个样子的
总结,生存分析图需要会画,包括细节的生存相关的概念。还有就是上面从FIG 9到FIG10的过渡,没有搞清,如何从9个基因过渡到FIG10的那些基因的。
最后友情宣传生信技能树
生物信息学“义诊”
生物信息学”拍卖会”
全国巡讲:R基础,Linux基础和RNA-seq实战演练 : 预告:12月28-30长沙站
广州珠江新城GEO数据挖掘滚动开班
DNA及RNA甲基化数据分析与课题设计