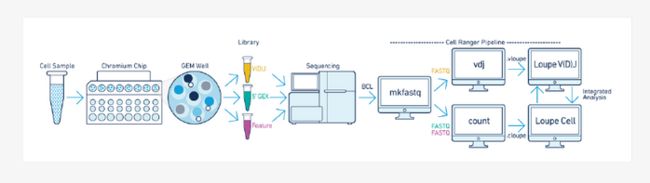
在做10X单细胞免疫组库分析的是往往是做一部分BCR、TCR做一部分5‘转录组,那么怎样才能把两者结合到一起呢?
今天我们尝试用我们的趁手工具做一下整合分析。
首先是下载数据,我们从10X官方的dataset中下载数据:https://support.10xgenomics.com/single-cell-vdj/datasets/3.1.0/vdj_v1_hs_pbmc3
在下载页面有关于这个样本的基本介绍,如这个数据集根据单细胞V(D)J试剂试剂盒使用指南和细胞表面蛋白特征条形码技术(CG000186),从标记的细胞中扩增出cDNA,生成5’基因表达、细胞表面蛋白、TCR富集和Ig富集文库。这里是不是应该把本文的题目改成:Seurat整合单细胞转录组与VDJ数据与膜蛋白数据啊。
读入数据:
rm(list =ls())
library(Seurat)
data <- Read10X(data.dir = "F:\\VDJ\\filtered_feature_bc_matrix")
10X data contains more than one type and is being returned as a list containing matrices of each type.
上来就报错:
seurat_object = CreateSeuratObject(counts = data)
Error in CreateAssayObject(counts = counts, min.cells = min.cells, min.features = min.features) :
No cell names (colnames) names present in the input matrix
>
原来我们的feature中不仅有基因表达还有膜蛋白的信息,分开来读好了:
seurat_object = CreateSeuratObject(counts = data$`Gene Expression`)
seurat_object[['Protein']] = CreateAssayObject(counts = data$`Antibody Capture`)
我们熟悉的seurat对象又回来了:
seurat_object
An object of class Seurat
33555 features across 7231 samples within 2 assays
Active assay: RNA (33538 features)
1 other assay present: Protein
注意这里是两个数据, RNA ,Protein:
当我们需要分析哪一部分的时候,用DefaultAssay(seurat_object)来指定,这里我们只分析RNA的数据啊,茂密的森林伸出两条路,我们也看一眼膜蛋白的数据吧:
seurat_object@assays$Protein
Assay data with 17 features for 7231 cells
First 10 features:
CD3-UCHT1-TotalC, CD19-HIB19-TotalC, CD45RA-HI100-TotalC, CD4-RPA-T4-TotalC, CD8a-RPA-T8-TotalC, CD14-M5E2-TotalC, CD16-3G8-TotalC, CD56-QA17A16-TotalC, CD25-BC96-TotalC,
CD45RO-UCHL1-TotalC
接着我们读入BCR的数据:
bcr <- read.csv("F:\\VDJ\\filtered_contig_annotations.csv")
bcr[1:4,]
barcode is_cell contig_id high_confidence length chain v_gene d_gene j_gene c_gene full_length productive cdr3
1 AAACCTGTCACTGGGC-1 True AAACCTGTCACTGGGC-1_contig_1 True 556 IGK IGKV1D-33 None IGKJ4 IGKC True True CQQYDNLPLTF
2 AAACCTGTCACTGGGC-1 True AAACCTGTCACTGGGC-1_contig_2 True 516 IGH IGHV4-59 None IGHJ5 IGHM True True CARGGNSGLDPW
3 AAACCTGTCACTGGGC-1 True AAACCTGTCACTGGGC-1_contig_3 True 569 IGK IGKV2D-30 None IGKJ1 IGKC True False CWGLLLHARYTLAWTF
4 AAACCTGTCAGGTAAA-1 True AAACCTGTCAGGTAAA-1_contig_1 True 548 IGK IGKV1-12 None IGKJ4 IGKC True True CQQANSFPLTF
cdr3_nt reads umis raw_clonotype_id raw_consensus_id
1 TGTCAACAGTATGATAATCTCCCGCTCACTTTC 7410 61 clonotype7 clonotype7_consensus_1
2 TGTGCGAGAGGCGGGAACAGTGGCTTAGACCCCTGG 1458 18 clonotype7 clonotype7_consensus_2
3 TGTTGGGGTTTATTACTGCATGCAAGGTACACACTGGCCTGGACGTTC 2045 17 clonotype7 None
4 TGTCAACAGGCTAACAGTTTCCCGCTCACTTTC 140 1 clonotype2 clonotype2_consensus_2
为了保持barcode一致,处理一下barcode的命名:
bcr$barcode <- gsub("-1", "", bcr$barcode)
bcr <- bcr[!duplicated(bcr$barcode), ]
names(bcr)[names(bcr) == "raw_clonotype_id"] <- "clonotype_id"
table(bcr$v_gene)
IGHV1-18 IGHV1-2 IGHV1-24 IGHV1-3 IGHV1-45 IGHV1-46 IGHV1-58 IGHV1-69 IGHV1-69D IGHV1-8 IGHV1/OR15-1 IGHV1/OR21-1 IGHV2-26 IGHV2-5 IGHV2-70
30 48 5 14 1 18 2 1 26 8 1 1 9 28 8
IGHV3-11 IGHV3-13 IGHV3-15 IGHV3-16 IGHV3-21 IGHV3-23 IGHV3-30 IGHV3-33 IGHV3-43 IGHV3-48 IGHV3-49 IGHV3-53 IGHV3-64 IGHV3-64D IGHV3-66
20 7 29 1 43 95 40 50 10 25 10 13 5 4 2
IGHV3-7 IGHV3-72 IGHV3-73 IGHV3-74 IGHV4-30-2 IGHV4-31 IGHV4-34 IGHV4-39 IGHV4-4 IGHV4-59 IGHV4-61 IGHV5-10-1 IGHV5-51 IGHV6-1 IGHV7-4-1
15 6 7 25 9 4 30 55 22 76 9 5 21 4 8
IGKV1-12 IGKV1-16 IGKV1-17 IGKV1-27 IGKV1-33 IGKV1-37 IGKV1-39 IGKV1-5 IGKV1-6 IGKV1-8 IGKV1-9 IGKV1D-13 IGKV1D-33 IGKV1D-37 IGKV1D-39
29 10 8 9 9 2 6 42 3 16 11 1 15 6 65
IGKV1D-43 IGKV1D-8 IGKV2-24 IGKV2-28 IGKV2-30 IGKV2D-26 IGKV2D-28 IGKV2D-29 IGKV2D-30 IGKV2D-40 IGKV3-11 IGKV3-15 IGKV3-20 IGKV3-7 IGKV3D-11
5 4 6 26 14 5 1 9 1 2 39 58 89 2 1
IGKV3D-15 IGKV3D-20 IGKV4-1 IGKV5-2 IGKV6-21 IGKV6D-21 IGLV1-40 IGLV1-44 IGLV1-47 IGLV1-51 IGLV10-54 IGLV2-11 IGLV2-14 IGLV2-18 IGLV2-23
9 2 60 8 2 1 31 37 26 24 3 24 60 1 20
IGLV2-8 IGLV3-1 IGLV3-10 IGLV3-12 IGLV3-16 IGLV3-19 IGLV3-21 IGLV3-25 IGLV3-27 IGLV3-9 IGLV4-3 IGLV4-60 IGLV4-69 IGLV5-37 IGLV5-39
20 35 15 1 2 15 27 16 4 4 2 5 15 7 1
IGLV5-45 IGLV5-48 IGLV5-52 IGLV6-57 IGLV7-43 IGLV7-46 IGLV8-61 IGLV9-49 None
9 1 2 14 5 10 15 9 173
>
读入克隆型信息:
clono <- read.csv("F:\\VDJ\\vdj_v1_hs_pbmc3_b_clonotypes.csv")
head(clono)
clonotype_id frequency proportion cdr3s_aa cdr3s_nt
1 clonotype1 7 0.009067358 IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
2 clonotype2 3 0.003886010 IGK:CQQANSFPLTF;IGK:CQQYDNWPPYTF IGK:TGTCAACAGGCTAACAGTTTCCCGCTCACTTTC;IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
3 clonotype5 2 0.002590674 IGL:CQARDSSTVVF IGL:TGTCAGGCGCGGGACAGCAGCACTGTGGTATTC
4 clonotype6 2 0.002590674 IGH:CARPGTTGTTGLKNW;IGK:CQQYNNWPLTF IGH:TGTGCGAGACCCGGTACAACTGGAACGACGGGTTTAAAAAACTGG;IGK:TGTCAGCAGTATAATAACTGGCCTCTCACCTTC
5 clonotype4 2 0.002590674 IGH:CAKSFFTGTGQFHYW;IGK:CQQYSTYSQTF IGH:TGTGCGAAATCATTTTTTACCGGGACAGGACAGTTTCACTATTGG;IGK:TGCCAACAGTATAGTACTTATTCTCAAACGTTC
6 clonotype3 2 0.002590674 IGL:CQSYDSSLSGVF IGL:TGCCAGTCCTATGACAGCAGCCTGAGTGGGGTGTTC
整合到seurat之中:
# Slap the AA sequences onto our original table by clonotype_id.
bcr <- merge(bcr, clono)
# Reorder so barcodes are first column and set them as rownames.
rownames(bcr) <- bcr[,2]
#tcr[,1] <- NULL
# Add to the Seurat object's metadata.
clono_seurat <- AddMetaData(object=seurat_object, metadata=bcr)
head([email protected])
orig.ident nCount_RNA nFeature_RNA nCount_Protein nFeature_Protein clonotype_id barcode is_cell contig_id high_confidence length chain v_gene d_gene j_gene c_gene full_length
AAACCTGAGATCTGAA SeuratProject 4386 1206 5092 17 NA
AAACCTGAGGAACTGC SeuratProject 6258 1723 6480 17 NA
AAACCTGAGGAGTCTG SeuratProject 4243 1404 3830 17 NA
AAACCTGAGGCTCTTA SeuratProject 5205 1622 3589 17 NA
AAACCTGAGTACGTTC SeuratProject 7098 2123 5250 17 NA
AAACCTGGTATCACCA SeuratProject 1095 604 3573 17 NA
productive cdr3 cdr3_nt reads umis raw_consensus_id frequency proportion cdr3s_aa cdr3s_nt
AAACCTGAGATCTGAA NA NA NA NA
AAACCTGAGGAACTGC NA NA NA NA
AAACCTGAGGAGTCTG NA NA NA NA
AAACCTGAGGCTCTTA NA NA NA NA
AAACCTGAGTACGTTC NA NA NA NA
AAACCTGGTATCACCA NA NA NA NA
为什么这么多NA?因为转录组中的细胞很多,而免疫细胞(BCR)只有一部分。
table(rownames([email protected]) %in% rownames(bcr))
FALSE TRUE
6469 762
> clono_seurat<- subset(clono_seurat,cells = rownames(bcr))
> head([email protected])
orig.ident nCount_RNA nFeature_RNA nCount_Protein nFeature_Protein clonotype_id barcode is_cell contig_id high_confidence length chain v_gene d_gene
CTTTGCGCAAGGACTG SeuratProject 1266 642 696 17 clonotype1 CTTTGCGCAAGGACTG True CTTTGCGCAAGGACTG-1_contig_1 True 479 IGK IGKV3-15 None
CAGCTAACACCGATAT SeuratProject 935 484 545 17 clonotype1 CAGCTAACACCGATAT True CAGCTAACACCGATAT-1_contig_1 True 549 IGK IGKV3-15 None
TATTACCGTCGCTTCT SeuratProject 1352 594 422 17 clonotype1 TATTACCGTCGCTTCT True TATTACCGTCGCTTCT-1_contig_1 True 565 IGK IGKV3-15 None
CTGAAGTGTGAAGGCT SeuratProject 1259 626 450 17 clonotype1 CTGAAGTGTGAAGGCT True CTGAAGTGTGAAGGCT-1_contig_1 True 554 IGK IGKV3-15 None
CCATGTCAGACCGGAT SeuratProject 1088 555 741 17 clonotype1 CCATGTCAGACCGGAT True CCATGTCAGACCGGAT-1_contig_1 True 556 IGK IGKV3-15 None
CGTTCTGAGCTCAACT SeuratProject 808 461 4896 17 clonotype1 CGTTCTGAGCTCAACT True CGTTCTGAGCTCAACT-1_contig_1 True 565 IGK IGKV3-15 None
j_gene c_gene full_length productive cdr3 cdr3_nt reads umis raw_consensus_id frequency proportion cdr3s_aa
CTTTGCGCAAGGACTG IGKJ2 IGKC True True CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 197 2 clonotype1_consensus_1 7 0.009067358 IGK:CQQYDNWPPYTF
CAGCTAACACCGATAT IGKJ2 IGKC True True CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 274 2 clonotype1_consensus_1 7 0.009067358 IGK:CQQYDNWPPYTF
TATTACCGTCGCTTCT IGKJ2 IGKC True True CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 299 2 clonotype1_consensus_1 7 0.009067358 IGK:CQQYDNWPPYTF
CTGAAGTGTGAAGGCT IGKJ2 IGKC True True CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 250 3 clonotype1_consensus_1 7 0.009067358 IGK:CQQYDNWPPYTF
CCATGTCAGACCGGAT IGKJ2 IGKC True True CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 324 2 clonotype1_consensus_1 7 0.009067358 IGK:CQQYDNWPPYTF
CGTTCTGAGCTCAACT IGKJ2 IGKC True True CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 187 3 clonotype1_consensus_1 7 0.009067358 IGK:CQQYDNWPPYTF
cdr3s_nt
CTTTGCGCAAGGACTG IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
CAGCTAACACCGATAT IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
TATTACCGTCGCTTCT IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
CTGAAGTGTGAAGGCT IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
CCATGTCAGACCGGAT IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
CGTTCTGAGCTCAACT IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT
seurat标准过程:
library(tidyverse)
clono_seurat %>% NormalizeData() %>% FindVariableFeatures() %>%
ScaleData(assay="RNA") %>%
RunPCA(npcs = 50,assay="RNA")%>%
FindNeighbors(assay="RNA")%>%
FindClusters(assay="RNA")%>%
RunUMAP(1:30) -> clono_seurat
head([email protected])
orig.ident nCount_RNA nFeature_RNA nCount_Protein nFeature_Protein clonotype_id barcode is_cell
CTTTGCGCAAGGACTG SeuratProject 1266 642 696 17 clonotype1 CTTTGCGCAAGGACTG True
CAGCTAACACCGATAT SeuratProject 935 484 545 17 clonotype1 CAGCTAACACCGATAT True
TATTACCGTCGCTTCT SeuratProject 1352 594 422 17 clonotype1 TATTACCGTCGCTTCT True
CTGAAGTGTGAAGGCT SeuratProject 1259 626 450 17 clonotype1 CTGAAGTGTGAAGGCT True
CCATGTCAGACCGGAT SeuratProject 1088 555 741 17 clonotype1 CCATGTCAGACCGGAT True
CGTTCTGAGCTCAACT SeuratProject 808 461 4896 17 clonotype1 CGTTCTGAGCTCAACT True
contig_id high_confidence length chain v_gene d_gene j_gene c_gene full_length productive
CTTTGCGCAAGGACTG CTTTGCGCAAGGACTG-1_contig_1 True 479 IGK IGKV3-15 None IGKJ2 IGKC True True
CAGCTAACACCGATAT CAGCTAACACCGATAT-1_contig_1 True 549 IGK IGKV3-15 None IGKJ2 IGKC True True
TATTACCGTCGCTTCT TATTACCGTCGCTTCT-1_contig_1 True 565 IGK IGKV3-15 None IGKJ2 IGKC True True
CTGAAGTGTGAAGGCT CTGAAGTGTGAAGGCT-1_contig_1 True 554 IGK IGKV3-15 None IGKJ2 IGKC True True
CCATGTCAGACCGGAT CCATGTCAGACCGGAT-1_contig_1 True 556 IGK IGKV3-15 None IGKJ2 IGKC True True
CGTTCTGAGCTCAACT CGTTCTGAGCTCAACT-1_contig_1 True 565 IGK IGKV3-15 None IGKJ2 IGKC True True
cdr3 cdr3_nt reads umis raw_consensus_id frequency proportion
CTTTGCGCAAGGACTG CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 197 2 clonotype1_consensus_1 7 0.009067358
CAGCTAACACCGATAT CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 274 2 clonotype1_consensus_1 7 0.009067358
TATTACCGTCGCTTCT CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 299 2 clonotype1_consensus_1 7 0.009067358
CTGAAGTGTGAAGGCT CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 250 3 clonotype1_consensus_1 7 0.009067358
CCATGTCAGACCGGAT CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 324 2 clonotype1_consensus_1 7 0.009067358
CGTTCTGAGCTCAACT CQQYDNWPPYTF TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 187 3 clonotype1_consensus_1 7 0.009067358
cdr3s_aa cdr3s_nt RNA_snn_res.0.8 seurat_clusters
CTTTGCGCAAGGACTG IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 5 5
CAGCTAACACCGATAT IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 5 5
TATTACCGTCGCTTCT IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 5 5
CTGAAGTGTGAAGGCT IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 5 5
CCATGTCAGACCGGAT IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 5 5
CGTTCTGAGCTCAACT IGK:CQQYDNWPPYTF IGK:TGTCAGCAGTATGATAACTGGCCTCCGTACACTTTT 5 5
p1 <- DimPlot(clono_seurat)
p2 <-DimPlot(clono_seurat,group.by = "chain")
p3 <-FeaturePlot(clono_seurat,features="frequency")
library(patchwork)
p1+p2+p3
library(sankeywheel)
sankeywheel(
from = [email protected]$chain, to = [email protected]$v_gene,
weight = [email protected]$nCount_Protein,#type = "sankey",
title = "sk",
subtitle = "chain & v_gene",
seriesName = "", width = "100%", height = "600px"
)
计算不同链的差异基因:
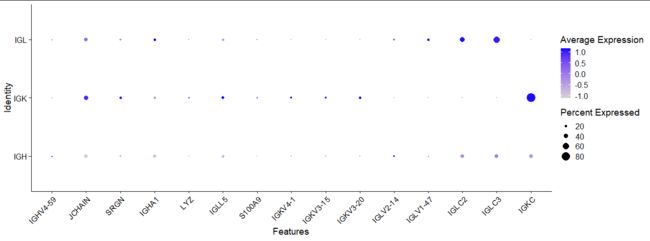
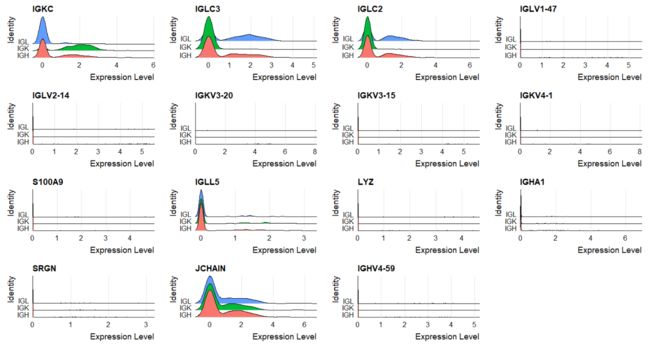
DT::datatable(FindMarkers(clono_seurat,ident.1 = 'IGK',group.by = "chain"))
mk<-FindMarkers(clono_seurat,ident.1 = 'IGK',group.by = "chain")
DotPlot(clono_seurat,features=rownames(mk),group.by = "chain") + RotatedAxis()
RidgePlot(clono_seurat,features=rownames(mk),group.by = "chain")
Tutorial: Integrating VDJ sequencing data with Seurat
Tutorial: Associating VDJ clonotyping data with scRNA-seq in Seurat