真菌和细菌高通量测序引物选择
最常用的真菌ITS1区引物
细菌内生菌测序引物
进行巢式PCR引物选择及反应条件
一些引物选择相关文献:
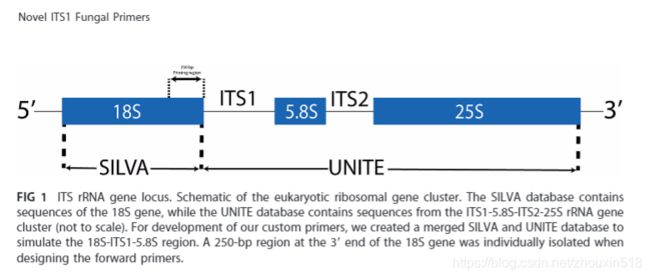
The fungal primers ITS5-1737-F (5′-GGA AGT AAA AGT CGT AAC AAG G-3′) and ITS2-2043-R (5′-GCT GCG TTC TTC ATC GAT GC-3′) with specific barcode were used to amplify the ITS1 regions of the fungal ITS rRNA genes
本文结论
During PCR amplification of full-length fungal ITS fragments, community composition may be severely distorted because of discrimination against taxa with long amplicons. Using primers in the 5.8S region to amplify the ITS2 region only, such distortion may be reduced. Quantitative relations between different templates are then reasonably well preserved both for common and rare taxa, even though the relative precision is lower for less frequent taxa. Furthermore, the new primers fITS7 and gITS7 return a more diverse amplicon community than the ITS1f primer, presumably due to a combination of better nucleotide matching to primer sites and reduced discrimination against long templates. The primers fITS9 and fITS7 were found to be more or less specific to fungi, whereas the gITS7 primer also amplified many plants (but not conifers). On the other hand, the gITS7 primer yielded the most diverse amplicon communities.
苹果内生菌文章真菌和细菌引物: ITS3/KYO2 and ITS4;515F and 806R
(Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence)
The fungal ITS2 region was amplified using the universal primers ITS3/KYO2 and ITS4 to amplify the ITS2 region of the ribosomal DNA [30]. The bacterial 16S region was amplified using the protocol described by Lundberg et al. [31], including the use of a pair of peptide-nucleic-acids (PNA) that were incorporated into the PCR amplification in order to reduce the generation of non-target chloroplast and mitochondrial amplicons. The universal 16S primer pair 515F and 806R was used to generate bacterial-derived 16S amplicons [31]. All primers were modified to include Illumina
adaptors (www.illumina.com). PCR reactions were conducted in a total volume of 25 μL containing 12.5 μL of KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA), 1.0 μL of each primer (10 μM), 2.5 μL of DNA template, and 8.0 μL nuclease-free water. The reactions were incubated in a T100 thermal cycler (Bio-Rad) for 3 min at 98 °C followed by 30 cycles of 30 s
at 95 °C, 30 s at 50 °C, and 30 s at 72 °C. All reactions ended with a final extension of 1 min at 72 °C. Nucleasefree water (QIAGEN, Valencia, CA, USA) replaced template DNA in negative controls. All amplicons and amplification mixtures including negative controls were sequenced on a MiSeq platform using V2 chemistry (Illumina, San Diego, CA, USA).
根系微生物介导磷压力文章V4 (515F–806R)
(Root microbiota drive direct integration of phosphate stress and immunity)
Three sets of index primers were used to amplify the V4 (515F–806R) region of the 16S rRNA gene of each sample.
甘蔗微生物组y引物: 515f and 806r; ITS9 and ITS4 primers to target the ITS2
(Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome)
Primer design. To access the bacterial and archaea communities, the V4 region of the 16S ribosomal gene was targeted using the 515f and 806r primers45 with modifed overhangs (Supplementary data). Te fungal community diversity and abundance was accessed using modifed ITS9 and ITS4 primers to target the ITS2 region32 (Supplementary data).