【机器学习】支持向量机(SVM)代码练习
本课程是中国大学慕课《机器学习》的“支持向量机”章节的课后代码。
课程地址:
https://www.icourse163.org/course/WZU-1464096179
课程完整代码:
https://github.com/fengdu78/WZU-machine-learning-course
代码修改并注释:黄海广,[email protected]
在本练习中,我们将使用支持向量机(SVM)来构建垃圾邮件分类器。我们将从一些简单的2D数据集开始使用SVM来查看它们的工作原理。然后,我们将对一组原始电子邮件进行一些预处理工作,并使用SVM在处理的电子邮件上构建分类器,以确定它们是否为垃圾邮件。
我们要做的第一件事是看一个简单的二维数据集,看看线性SVM如何对数据集进行不同的C值(类似于线性/逻辑回归中的正则化项)。
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sbimport warnings
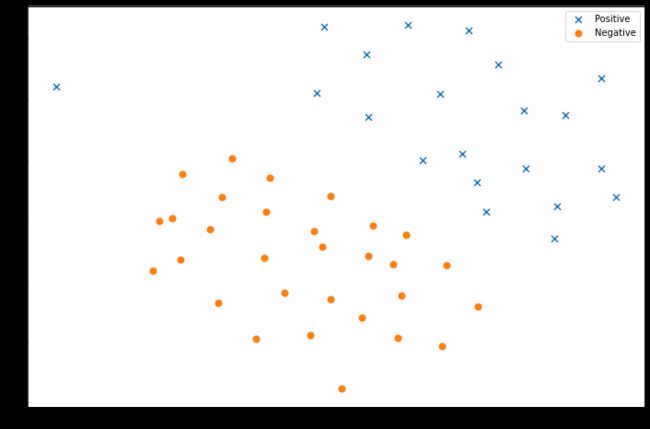
warnings.simplefilter("ignore")我们将其用散点图表示,其中类标签由符号表示(+表示正类,o表示负类)。
data1 = pd.read_csv('data/svmdata1.csv')data1.head()| X1 | X2 | y | |
|---|---|---|---|
| 0 | 1.9643 | 4.5957 | 1 |
| 1 | 2.2753 | 3.8589 | 1 |
| 2 | 2.9781 | 4.5651 | 1 |
| 3 | 2.9320 | 3.5519 | 1 |
| 4 | 3.5772 | 2.8560 | 1 |
positive = data1[data1['y'].isin([1])]
negative = data1[data1['y'].isin([0])]
fig, ax = plt.subplots(figsize=(12, 8))
ax.scatter(positive['X1'], positive['X2'], s=50, marker='x', label='Positive')
ax.scatter(negative['X1'], negative['X2'], s=50, marker='o', label='Negative')
ax.legend()
plt.show()请注意,还有一个异常的正例在其他样本之外。 这些类仍然是线性分离的,但它非常紧凑。我们要训练线性支持向量机来学习类边界。在这个练习中,我们没有从头开始执行SVM的任务,所以我要用scikit-learn。
from sklearn import svm
svc = svm.LinearSVC(C=1, loss='hinge', max_iter=1000)
svcLinearSVC(C=1, loss='hinge')首先,我们使用 C=1 看下结果如何。
svc.fit(data1[['X1', 'X2']], data1['y'])
svc.score(data1[['X1', 'X2']], data1['y'])0.9803921568627451其次,让我们看看如果C的值越大,会发生什么
svc2 = svm.LinearSVC(C=100, loss='hinge', max_iter=1000)
svc2.fit(data1[['X1', 'X2']], data1['y'])
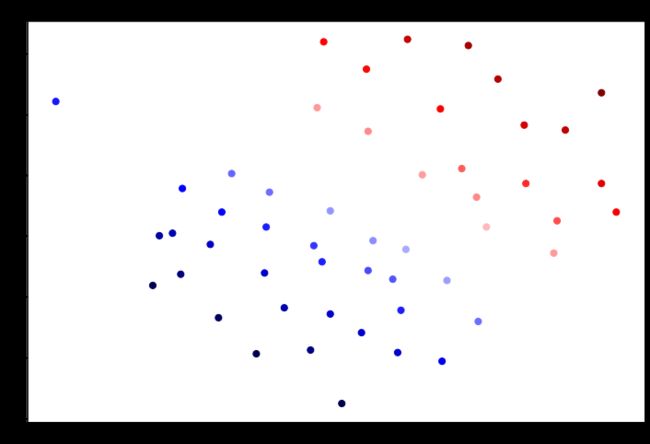
svc2.score(data1[['X1', 'X2']], data1['y'])0.9411764705882353这次我们得到了训练数据的完美分类,但是通过增加C的值,我们创建了一个不再适合数据的决策边界。我们可以通过查看每个类别预测的置信水平来看出这一点,这是该点与超平面距离的函数。
data1['SVM 1 Confidence'] = svc.decision_function(data1[['X1', 'X2']])
fig, ax = plt.subplots(figsize=(12, 8))
ax.scatter(data1['X1'],
data1['X2'],
s=50,
c=data1['SVM 1 Confidence'],
cmap='seismic')
ax.set_title('SVM (C=1) Decision Confidence')
plt.show()data1['SVM 2 Confidence'] = svc2.decision_function(data1[['X1', 'X2']])
fig, ax = plt.subplots(figsize=(12,8))
ax.scatter(data1['X1'], data1['X2'], s=50, c=data1['SVM 2 Confidence'], cmap='seismic')
ax.set_title('SVM (C=100) Decision Confidence')
plt.show()可以看看靠近边界的点的颜色,区别是有点微妙。如果您在练习文本中,则会出现绘图,其中决策边界在图上显示为一条线,有助于使差异更清晰。
现在我们将从线性SVM转移到能够使用内核进行非线性分类的SVM。我们首先负责实现一个高斯核函数。虽然scikit-learn具有内置的高斯内核,但为了实现更清楚,我们将从头开始实现。
def gaussian_kernel(x1, x2, sigma):
return np.exp(-(np.sum((x1 - x2)**2) / (2 * (sigma**2))))x1 = np.array([1.0, 2.0, 1.0])
x2 = np.array([0.0, 4.0, -1.0])
sigma = 2
gaussian_kernel(x1, x2, sigma)0.32465246735834974该结果与练习中的预期值相符。接下来,我们将检查另一个数据集,这次用非线性决策边界。
data2 = pd.read_csv('data/svmdata2.csv')data2.head()| X1 | X2 | y | |
|---|---|---|---|
| 0 | 0.107143 | 0.603070 | 1 |
| 1 | 0.093318 | 0.649854 | 1 |
| 2 | 0.097926 | 0.705409 | 1 |
| 3 | 0.155530 | 0.784357 | 1 |
| 4 | 0.210829 | 0.866228 | 1 |
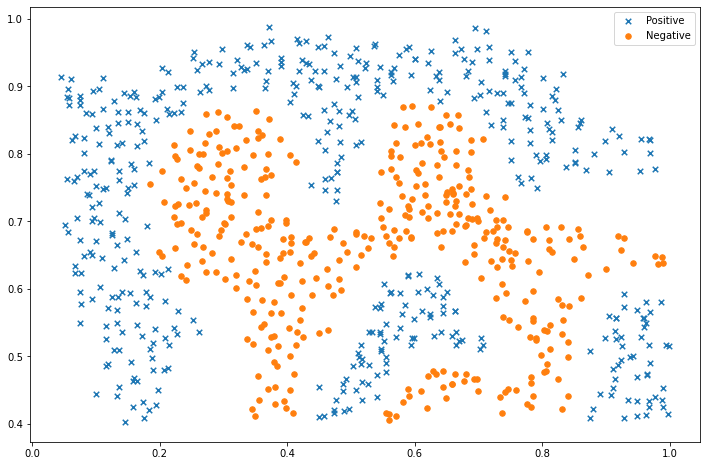
positive = data2[data2['y'].isin([1])]
negative = data2[data2['y'].isin([0])]
fig, ax = plt.subplots(figsize=(12, 8))
ax.scatter(positive['X1'], positive['X2'], s=30, marker='x', label='Positive')
ax.scatter(negative['X1'], negative['X2'], s=30, marker='o', label='Negative')
ax.legend()
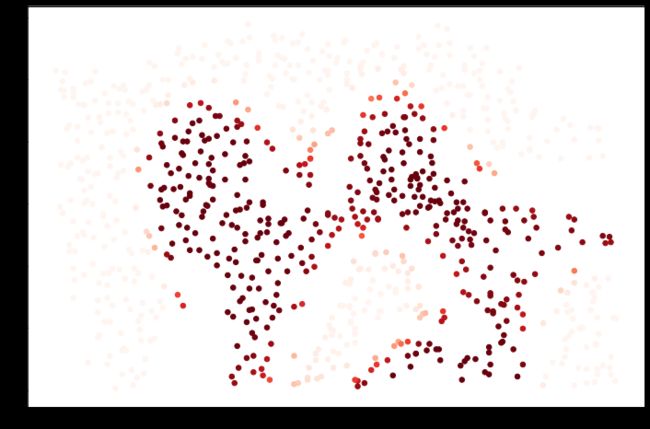
plt.show()对于该数据集,我们将使用内置的RBF内核构建支持向量机分类器,并检查其对训练数据的准确性。为了可视化决策边界,这一次我们将根据实例具有负类标签的预测概率来对点做阴影。从结果可以看出,它们大部分是正确的。
svc = svm.SVC(C=100, gamma=10, probability=True)
svcSVC(C=100, gamma=10, probability=True)svc.fit(data2[['X1', 'X2']], data2['y'])
svc.score(data2[['X1', 'X2']], data2['y'])0.9698725376593279data2['Probability'] = svc.predict_proba(data2[['X1', 'X2']])[:, 0]
fig, ax = plt.subplots(figsize=(12, 8))
ax.scatter(data2['X1'], data2['X2'], s=30, c=data2['Probability'], cmap='Reds')
plt.show()对于第三个数据集,我们给出了训练和验证集,并且基于验证集性能为SVM模型找到最优超参数。虽然我们可以使用scikit-learn的内置网格搜索来做到这一点,但是本着遵循练习的目的,我们将从头开始实现一个简单的网格搜索。
data3=pd.read_csv('data/svmdata3.csv')
data3val=pd.read_csv('data/svmdata3val.csv')X = data3[['X1','X2']]
Xval = data3val[['X1','X2']]
y = data3['y'].ravel()
yval = data3val['yval'].ravel()C_values = [0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100]
gamma_values = [0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100]
best_score = 0
best_params = {'C': None, 'gamma': None}
for C in C_values:
for gamma in gamma_values:
svc = svm.SVC(C=C, gamma=gamma)
svc.fit(X, y)
score = svc.score(Xval, yval)
if score > best_score:
best_score = score
best_params['C'] = C
best_params['gamma'] = gamma
best_score, best_params(0.965, {'C': 0.3, 'gamma': 100})大间隔分类器
from sklearn.svm import SVC
from sklearn import datasets
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
iris = datasets.load_iris()
X = iris["data"][:, (2, 3)] # petal length, petal width
y = iris["target"]
setosa_or_versicolor = (y == 0) | (y == 1)
X = X[setosa_or_versicolor]
y = y[setosa_or_versicolor]
# SVM Classifier model
svm_clf = SVC(kernel="linear", C=float("inf"))
svm_clf.fit(X, y)SVC(C=inf, kernel='linear')# Bad models
x0 = np.linspace(0, 5.5, 200)
pred_1 = 5 * x0 - 20
pred_2 = x0 - 1.8
pred_3 = 0.1 * x0 + 0.5def plot_svc_decision_boundary(svm_clf, xmin, xmax):
w = svm_clf.coef_[0]
b = svm_clf.intercept_[0]
# At the decision boundary, w0*x0 + w1*x1 + b = 0
# => x1 = -w0/w1 * x0 - b/w1
x0 = np.linspace(xmin, xmax, 200)
decision_boundary = -w[0]/w[1] * x0 - b/w[1]
margin = 1/w[1]
gutter_up = decision_boundary + margin
gutter_down = decision_boundary - margin
svs = svm_clf.support_vectors_
plt.scatter(svs[:, 0], svs[:, 1], s=180, facecolors='#FFAAAA')
plt.plot(x0, decision_boundary, "k-", linewidth=2)
plt.plot(x0, gutter_up, "k--", linewidth=2)
plt.plot(x0, gutter_down, "k--", linewidth=2)plt.figure(figsize=(12, 2.7))
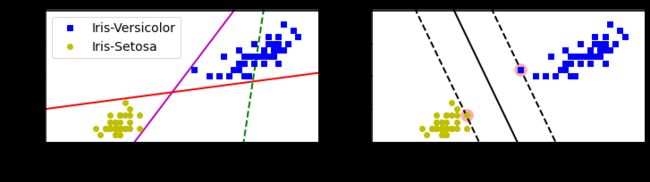
plt.subplot(121)
plt.plot(x0, pred_1, "g--", linewidth=2)
plt.plot(x0, pred_2, "m-", linewidth=2)
plt.plot(x0, pred_3, "r-", linewidth=2)
plt.plot(X[:, 0][y == 1], X[:, 1][y == 1], "bs", label="Iris-Versicolor")
plt.plot(X[:, 0][y == 0], X[:, 1][y == 0], "yo", label="Iris-Setosa")
plt.xlabel("Petal length", fontsize=14)
plt.ylabel("Petal width", fontsize=14)
plt.legend(loc="upper left", fontsize=14)
plt.axis([0, 5.5, 0, 2])
plt.subplot(122)
plot_svc_decision_boundary(svm_clf, 0, 5.5)
plt.plot(X[:, 0][y == 1], X[:, 1][y == 1], "bs")
plt.plot(X[:, 0][y == 0], X[:, 1][y == 0], "yo")
plt.xlabel("Petal length", fontsize=14)
plt.axis([0, 5.5, 0, 2])
plt.show()特征缩放的敏感性
Xs = np.array([[1, 50], [5, 20], [3, 80], [5, 60]]).astype(np.float64)
ys = np.array([0, 0, 1, 1])
svm_clf = SVC(kernel="linear", C=100)
svm_clf.fit(Xs, ys)
plt.figure(figsize=(12, 3.2))
plt.subplot(121)
plt.plot(Xs[:, 0][ys == 1], Xs[:, 1][ys == 1], "bo")
plt.plot(Xs[:, 0][ys == 0], Xs[:, 1][ys == 0], "ms")
plot_svc_decision_boundary(svm_clf, 0, 6)
plt.xlabel("$x_0$", fontsize=20)
plt.ylabel("$x_1$ ", fontsize=20, rotation=0)
plt.title("Unscaled", fontsize=16)
plt.axis([0, 6, 0, 90])
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_scaled = scaler.fit_transform(Xs)
svm_clf.fit(X_scaled, ys)
plt.subplot(122)
plt.plot(X_scaled[:, 0][ys == 1], X_scaled[:, 1][ys == 1], "bo")
plt.plot(X_scaled[:, 0][ys == 0], X_scaled[:, 1][ys == 0], "ms")
plot_svc_decision_boundary(svm_clf, -2, 2)
plt.xlabel("$x_0$", fontsize=20)
plt.title("Scaled", fontsize=16)
plt.axis([-2, 2, -2, 2])
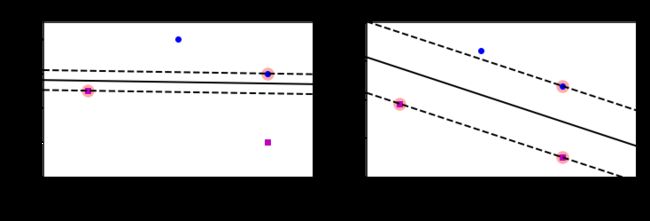
plt.show()硬间隔和软间隔分类
X_outliers = np.array([[3.4, 1.3], [3.2, 0.8]])
y_outliers = np.array([0, 0])
Xo1 = np.concatenate([X, X_outliers[:1]], axis=0)
yo1 = np.concatenate([y, y_outliers[:1]], axis=0)
Xo2 = np.concatenate([X, X_outliers[1:]], axis=0)
yo2 = np.concatenate([y, y_outliers[1:]], axis=0)
svm_clf2 = SVC(kernel="linear", C=10**9)
svm_clf2.fit(Xo2, yo2)
plt.figure(figsize=(12, 2.7))
plt.subplot(121)
plt.plot(Xo1[:, 0][yo1 == 1], Xo1[:, 1][yo1 == 1], "bs")
plt.plot(Xo1[:, 0][yo1 == 0], Xo1[:, 1][yo1 == 0], "yo")
plt.text(0.3, 1.0, "Impossible!", fontsize=24, color="red")
plt.xlabel("Petal length", fontsize=14)
plt.ylabel("Petal width", fontsize=14)
plt.annotate(
"Outlier",
xy=(X_outliers[0][0], X_outliers[0][1]),
xytext=(2.5, 1.7),
ha="center",
arrowprops=dict(facecolor='black', shrink=0.1),
fontsize=16,
)
plt.axis([0, 5.5, 0, 2])
plt.subplot(122)
plt.plot(Xo2[:, 0][yo2 == 1], Xo2[:, 1][yo2 == 1], "bs")
plt.plot(Xo2[:, 0][yo2 == 0], Xo2[:, 1][yo2 == 0], "yo")
plot_svc_decision_boundary(svm_clf2, 0, 5.5)
plt.xlabel("Petal length", fontsize=14)
plt.annotate(
"Outlier",
xy=(X_outliers[1][0], X_outliers[1][1]),
xytext=(3.2, 0.08),
ha="center",
arrowprops=dict(facecolor='black', shrink=0.1),
fontsize=16,
)
plt.axis([0, 5.5, 0, 2])
plt.show()from sklearn.pipeline import Pipelinefrom sklearn.datasets import make_moons
X, y = make_moons(n_samples=100, noise=0.15, random_state=42)def plot_predictions(clf, axes):
x0s = np.linspace(axes[0], axes[1], 100)
x1s = np.linspace(axes[2], axes[3], 100)
x0, x1 = np.meshgrid(x0s, x1s)
X = np.c_[x0.ravel(), x1.ravel()]
y_pred = clf.predict(X).reshape(x0.shape)
y_decision = clf.decision_function(X).reshape(x0.shape)
plt.contourf(x0, x1, y_pred, cmap=plt.cm.brg, alpha=0.2)
plt.contourf(x0, x1, y_decision, cmap=plt.cm.brg, alpha=0.1)def plot_dataset(X, y, axes):
plt.plot(X[:, 0][y==0], X[:, 1][y==0], "bs")
plt.plot(X[:, 0][y==1], X[:, 1][y==1], "g^")
plt.axis(axes)
plt.grid(True, which='both')
plt.xlabel(r"$x_1$", fontsize=20)
plt.ylabel(r"$x_2$", fontsize=20, rotation=0)from sklearn.svm import SVC
gamma1, gamma2 = 0.1, 5
C1, C2 = 0.001, 1000
hyperparams = (gamma1, C1), (gamma1, C2), (gamma2, C1), (gamma2, C2)
svm_clfs = []
for gamma, C in hyperparams:
rbf_kernel_svm_clf = Pipeline([("scaler", StandardScaler()),
("svm_clf",
SVC(kernel="rbf", gamma=gamma, C=C))])
rbf_kernel_svm_clf.fit(X, y)
svm_clfs.append(rbf_kernel_svm_clf)
plt.figure(figsize=(12, 7))
for i, svm_clf in enumerate(svm_clfs):
plt.subplot(221 + i)
plot_predictions(svm_clf, [-1.5, 2.5, -1, 1.5])
plot_dataset(X, y, [-1.5, 2.5, -1, 1.5])
gamma, C = hyperparams[i]
plt.title(r"$\gamma = {}, C = {}$".format(gamma, C), fontsize=12)
plt.show()svm推导
分离超平面:
点到直线距离:
为2-范数:
直线为超平面,样本可表示为:
margin:
函数间隔:
几何间隔:,当数据被正确分类时,几何间隔就是点到超平面的距离
为了求几何间隔最大,SVM基本问题可以转化为求解:(为几何间隔,(为函数间隔)
分类点几何间隔最大,同时被正确分类。但这个方程并非凸函数求解,所以要先①将方程转化为凸函数,②用拉格朗日乘子法和KKT条件求解对偶问题。
①转化为凸函数:
先令,方便计算(参照衡量,不影响评价结果)
再将转化成求解凸函数,1/2是为了求导之后方便计算。
②用拉格朗日乘子法和KKT条件求解最优值:
整合成:
推导:
根据KKT条件:
带入
再把max问题转成min问题:
以上为SVM对偶问题的对偶形式
kernel
在低维空间计算获得高维空间的计算结果,也就是说计算结果满足高维(满足高维,才能说明高维下线性可分)。
soft margin & slack variable
引入松弛变量,对应数据点允许偏离的functional margin 的量。
目标函数:
对偶问题:
Sequential Minimal Optimization
首先定义特征到结果的输出函数:.
因为
有
import numpy as np
import pandas as pd
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
%matplotlib inline# data
def create_data():
iris = load_iris()
df = pd.DataFrame(iris.data, columns=iris.feature_names)
df['label'] = iris.target
df.columns = ['sepal length', 'sepal width', 'petal length', 'petal width', 'label']
data = np.array(df.iloc[:100, [0, 1, -1]])
for i in range(len(data)):
if data[i,-1] == 0:
data[i,-1] = -1
# print(data)
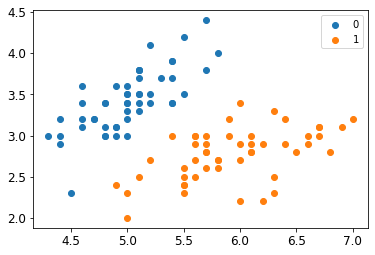
return data[:,:2], data[:,-1]X, y = create_data()
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)plt.scatter(X[:50,0],X[:50,1], label='0')
plt.scatter(X[50:,0],X[50:,1], label='1')
plt.legend()class SVM:
def __init__(self, max_iter=100, kernel='linear'):
self.max_iter = max_iter
self._kernel = kernel
def init_args(self, features, labels):
self.m, self.n = features.shape
self.X = features
self.Y = labels
self.b = 0.0
# 将Ei保存在一个列表里
self.alpha = np.ones(self.m)
self.E = [self._E(i) for i in range(self.m)]
# 松弛变量
self.C = 1.0
def _KKT(self, i):
y_g = self._g(i) * self.Y[i]
if self.alpha[i] == 0:
return y_g >= 1
elif 0 < self.alpha[i] < self.C:
return y_g == 1
else:
return y_g <= 1
# g(x)预测值,输入xi(X[i])
def _g(self, i):
r = self.b
for j in range(self.m):
r += self.alpha[j] * self.Y[j] * self.kernel(self.X[i], self.X[j])
return r
# 核函数
def kernel(self, x1, x2):
if self._kernel == 'linear':
return sum([x1[k] * x2[k] for k in range(self.n)])
elif self._kernel == 'poly':
return (sum([x1[k] * x2[k] for k in range(self.n)]) + 1)**2
return 0
# E(x)为g(x)对输入x的预测值和y的差
def _E(self, i):
return self._g(i) - self.Y[i]
def _init_alpha(self):
# 外层循环首先遍历所有满足0= 0:
j = min(range(self.m), key=lambda x: self.E[x])
else:
j = max(range(self.m), key=lambda x: self.E[x])
return i, j
def _compare(self, _alpha, L, H):
if _alpha > H:
return H
elif _alpha < L:
return L
else:
return _alpha
def fit(self, features, labels):
self.init_args(features, labels)
for t in range(self.max_iter):
# train
i1, i2 = self._init_alpha()
# 边界
if self.Y[i1] == self.Y[i2]:
L = max(0, self.alpha[i1] + self.alpha[i2] - self.C)
H = min(self.C, self.alpha[i1] + self.alpha[i2])
else:
L = max(0, self.alpha[i2] - self.alpha[i1])
H = min(self.C, self.C + self.alpha[i2] - self.alpha[i1])
E1 = self.E[i1]
E2 = self.E[i2]
# eta=K11+K22-2K12
eta = self.kernel(self.X[i1], self.X[i1]) + self.kernel(
self.X[i2],
self.X[i2]) - 2 * self.kernel(self.X[i1], self.X[i2])
if eta <= 0:
# print('eta <= 0')
continue
alpha2_new_unc = self.alpha[i2] + self.Y[i2] * (
E1 - E2) / eta #此处有修改,根据书上应该是E1 - E2,书上130-131页
alpha2_new = self._compare(alpha2_new_unc, L, H)
alpha1_new = self.alpha[i1] + self.Y[i1] * self.Y[i2] * (
self.alpha[i2] - alpha2_new)
b1_new = -E1 - self.Y[i1] * self.kernel(self.X[i1], self.X[i1]) * (
alpha1_new - self.alpha[i1]) - self.Y[i2] * self.kernel(
self.X[i2],
self.X[i1]) * (alpha2_new - self.alpha[i2]) + self.b
b2_new = -E2 - self.Y[i1] * self.kernel(self.X[i1], self.X[i2]) * (
alpha1_new - self.alpha[i1]) - self.Y[i2] * self.kernel(
self.X[i2],
self.X[i2]) * (alpha2_new - self.alpha[i2]) + self.b
if 0 < alpha1_new < self.C:
b_new = b1_new
elif 0 < alpha2_new < self.C:
b_new = b2_new
else:
# 选择中点
b_new = (b1_new + b2_new) / 2
# 更新参数
self.alpha[i1] = alpha1_new
self.alpha[i2] = alpha2_new
self.b = b_new
self.E[i1] = self._E(i1)
self.E[i2] = self._E(i2)
return 'train done!'
def predict(self, data):
r = self.b
for i in range(self.m):
r += self.alpha[i] * self.Y[i] * self.kernel(data, self.X[i])
return 1 if r > 0 else -1
def score(self, X_test, y_test):
right_count = 0
for i in range(len(X_test)):
result = self.predict(X_test[i])
if result == y_test[i]:
right_count += 1
return right_count / len(X_test)
def _weight(self):
# linear model
yx = self.Y.reshape(-1, 1) * self.X
self.w = np.dot(yx.T, self.alpha)
return self.w svm = SVM(max_iter=100)
svm.fit(X_train, y_train)'train done!'svm.score(X_test, y_test)0.6参考
Prof. Andrew Ng. Machine Learning. Stanford University
李航,《统计学习方法》,清华大学出版社