临床数据据清洗 临床表型数据 phenodata phe
对表型数据框进行去冗余 phe 表型信息提取 临床信息提取 自建函数提取 种方法数据框里面进行取子集操作,坐标、列名和逻辑判断每列满足某个要求每行满足某个要求按条件筛选数据库dataframe
https://www.r-bloggers.com/2021/04/tidyverse-in-r-complete-tutorial/
library(tidyverse)
library(nycflights13)
data(flights)
flights %>%
select(origin,everything())
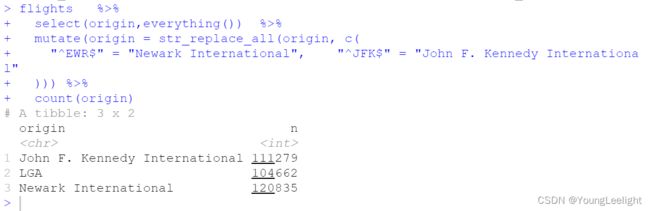
> flights %>%
+ select(origin,everything()) %>%
+ mutate(origin = str_replace_all(origin, c(
+ "^EWR$" = "Newark International", "^JFK$" = "John F. Kennedy International"
+ )))
pheno_data %>%
select(Group,everything()) %>%
mutate(Group2 = str_replace_all(Group, c(
"^abdomen_sepsis" = "Newark International",
"^JFK$" = "John F. Kennedy International"
))) %>% mutate(Group3=str_detect(Group2,"^ARDS_moderate"))
必须多加个mutate才可以 相当于mute把Group2的内容向量化 vectorized

flights %>%
select(origin,everything()) %>%
mutate(origin = str_replace_all(origin, c(
"^EWR$" = "Newark International", "^JFK$" = "John F. Kennedy International"
))) %>%
count(origin)
pheno_data=openxlsx::read.xlsx("G:/raw_material for omics_chaojie/Sepsis/sepsis和ARDS队列-患者信息.xlsx",
sheet=7)
head(pheno_data)
#制作wgcna的表型信息
#自建函数从字符串提取想要的字符,如果存在,就变成1,如果不存在则变成0
strsplit("abdomen_sepsis/ pneumonia_abdomen_sepsis/ abdomen_sepsis_Sshock",split = "/")
#r语言去除字符串两端多余空格
trimws(c(" abdo " ,' n_sepsi ',"s/ pneumonia"), which = c("both", "left", "right"), whitespace = "[ \t\r\n]")
any(c(1,3,4)==5)
get_mydesired_string=function(x,strings){
#strings="abdomen_sepsis/ abdomen/ sepsis/ ab"
#x="ab"
elements=strsplit(strings,split = "/")[[1]]
#r语言去除字符串两端多余空格 两边空格
elements=trimws(elements, which = c("both", "left", "right"), whitespace = "[ \t\r\n]")
if(any(which(elements==x))==TRUE)
{ myelements=elements[which(elements==x)]
myelements=1} else { myelements=0}
return(myelements)
}
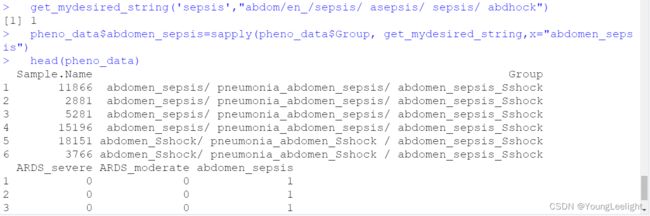
get_mydesired_string('sepsis',"abdom/en_/sepsis/ asepsis/ sepsis/ abdhock")
head(pheno_data)
pheno_data$'abdomen_sepsis'
str(pheno_data)
lapply(pheno_data$Group, get_mydesired_string,x="abdomen_sepsis")
sapply(pheno_data$Group, get_mydesired_string,x="abdomen_sepsis")
#以逗号为分隔取第几个元素
strsplit("15196、15197",split="、")
get_mydesired_element=function(location, strings){
#location=2
# strings="15196、15197"
elements=strsplit(strings,split="、")[[1]][location]
#去除空格
elements=elements=trimws(elements, which = c("both", "left", "right"), whitespace = "[ \t\r\n]")
return(elements)
}
#去掉括号及括号里的内容
{
gsub("\\(.*\\)","","afd90s(fsd)"); gsub("\\(.*\\)","","afd90sfsd)(,");gsub("\\(.*\\)","","afd90sfs")
sapply(colnames(lncrna),function(x){gsub(pattern = "\\(.*\\)","",x)})
gsub("\\(.*\\)","",colnames(lncrna))
#单纯去掉去掉中括号
gsub("\\[|\\]","",colnames(lncrna))
#单纯去掉括号
gsub("\\(|\\)","",colnames(lncrna))
}
#单纯去掉括号 和中括号
colnames(lncrna)= gsub("\\(|\\)","",colnames(lncrna))
colnames(lncrna)= gsub("\\[|\\]","",colnames(lncrna))
colnames(lncrna)