机器学习-聚类算法-02
文章目录
-
-
- 聚类算法实践
-
- 1. Kmeans
- 2. 决策边界
- 3. 算法流程
- 4. 不稳定的结果
- 5. 评估方法
- 6. 找到最佳簇数
- 7. 轮廓系数
- 8. Kmeans存在的问题
- 9. 图像分割实例
- 10. 半监督学习
- 11. DBSCAN算法
-
聚类算法实践
1. Kmeans
-
导包操作
导入numpy、matplotlib工具包,以及画图操作的相关参数设置,最后导入的包是防止环境出现警告
import numpy as np
import os
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
plt.rcParams['axes.labelsize'] = 14
plt.rcParams['xtick.labelsize'] = 12
plt.rcParams['ytick.labelsize'] = 12
import warnings
warnings.filterwarnings('ignore')
np.random.seed(42)
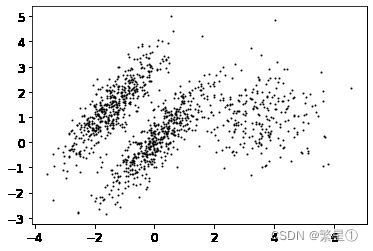
- 绘制中心点并设置发散程度
# 导入绘制类别的工具包
from sklearn.datasets import make_blobs
# 指定五个中心点
blob_centers = np.array(
[
[0.2,2.3],
[-1.5,2.3],
[-2.8,1.8],
[-2.8,2.8],
[-2.8,1.3]
]
)
# 五个点对应的发散程度,应当设置的小一点
blob_std = np.array([0.4,0.3,0.1,0.1,0.1])
- 设置样本
# 参数n_samples:样本个数,centers:样本中心点,cluster_std:以中心点为圆心,向周围发散的程度
X,y = make_blobs(n_samples = 2000,centers=blob_centers,cluster_std=blob_std,random_state=7)
- 画图展示
def plot_clusters(X,y = None):
plt.scatter(X[:,0],X[:,1],c=y,s=1)
plt.xlabel("$x_1$",fontsize=14)
plt.ylabel("$x_2$",fontsize=14,rotation=0)
plt.figure(figsize=(8,4))
# 无监督学习,传递参数的时候只传递X值
plot_clusters(X)
plt.show()
2. 决策边界
- 假定
k=5进行分类
from sklearn.cluster import KMeans
k = 5
kmeans = KMeans(n_clusters = 5,random_state = 42)
# 计算聚类中心并预测每个样本的聚类索引
# fit_predict(X)与kmeans.labels_得到的结果是一致的
y_pred = kmeans.fit_predict(X)
- 画图展示
# 把当前的数据进行展示
def plot_data(X):
plt.plot(X[:, 0], X[:, 1], 'k.', markersize=2)
# 绘制中心点
def plot_centroids(centroids, weights=None, circle_color='w', cross_color='k'):
if weights is not None:
centroids = centroids[weights > weights.max() / 10]
plt.scatter(centroids[:, 0], centroids[:, 1],
marker='o', s=30, linewidths=8,
color=circle_color, zorder=10, alpha=0.9)
plt.scatter(centroids[:, 0], centroids[:, 1],
marker='x', s=50, linewidths=5,
color=cross_color, zorder=11, alpha=1)
# 绘制等高线
def plot_decision_boundaries(clusterer, X, resolution=1000, show_centroids=True,
show_xlabels=True, show_ylabels=True):
mins = X.min(axis=0) - 0.1
maxs = X.max(axis=0) + 0.1
# 棋盘操作
xx, yy = np.meshgrid(np.linspace(mins[0], maxs[0], resolution),
np.linspace(mins[1], maxs[1], resolution))
# 预测结果值 合成棋盘
Z = clusterer.predict(np.c_[xx.ravel(), yy.ravel()])
# 预测结果值与棋盘一致
Z = Z.reshape(xx.shape)
# 绘制等高线,并绘制区域颜色
plt.contourf(Z, extent=(mins[0], maxs[0], mins[1], maxs[1]),
cmap="Pastel2")
plt.contour(Z, extent=(mins[0], maxs[0], mins[1], maxs[1]),
linewidths=1, colors='k')
plot_data(X)
if show_centroids:
plot_centroids(clusterer.cluster_centers_)
if show_xlabels:
plt.xlabel("$x_1$", fontsize=14)
else:
plt.tick_params(labelbottom='off')
if show_ylabels:
plt.ylabel("$x_2$", fontsize=14, rotation=0)
else:
plt.tick_params(labelleft='off')
# 调用函数开始绘图
plt.figure(figsize = (8,4))
plot_decision_boundaries(kmeans,X)
plt.show()
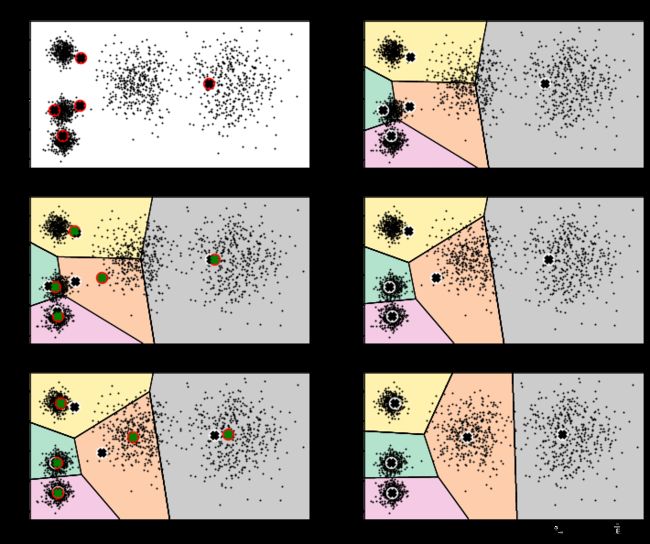
3. 算法流程
就是看程序每一步的优化结果
- 设置优化步数
# max_iter设置1/2/3相当于三步
kmean_iter1 = KMeans(n_clusters=5,init='random',n_init=1,max_iter=1,random_state=1)
kmean_iter2 = KMeans(n_clusters=5,init='random',n_init=1,max_iter=2,random_state=1)
kmean_iter3 = KMeans(n_clusters=5,init='random',n_init=1,max_iter=3,random_state=1)
kmean_iter1.fit(X)
kmean_iter2.fit(X)
kmean_iter3.fit(X)
- 画图展示
plt.figure(figsize=(12,10))
plt.subplot(321)
# 绘制初始点
plot_data(X)
# 绘制中心点
plot_centroids(kmean_iter1.cluster_centers_, circle_color='r', cross_color='k')
plt.title('Update cluster_centers')
plt.subplot(322)
plot_decision_boundaries(kmean_iter1, X,show_xlabels=False, show_ylabels=False)
plt.title('Iter1')
plt.subplot(323)
plot_decision_boundaries(kmean_iter1, X,show_xlabels=False, show_ylabels=False)
plot_centroids(kmean_iter2.cluster_centers_,circle_color='r',cross_color='g')
plt.title('Change1')
plt.subplot(324)
plot_decision_boundaries(kmean_iter2, X,show_xlabels=False, show_ylabels=False)
plt.title('Iter2')
plt.subplot(325)
plot_decision_boundaries(kmean_iter2, X,show_xlabels=False, show_ylabels=False)
plot_centroids(kmean_iter3.cluster_centers_,circle_color='r',cross_color='g')
plt.title('Change2')
plt.subplot(326)
plot_decision_boundaries(kmean_iter3, X,show_xlabels=False, show_ylabels=False)
plt.title('Iter3')
- 效果展示
图1为最开始的数据和生成的随机点分布;图2开始进行第一次迭代;图3是进行第二次迭代的过程,其中,黑白点代表上一次中心点的位置,红绿点代表本次迭代后中心点的更新;图4是第二次迭代后的中心点的位置;图5和图6分别是第三次迭代过程和结果中心点的分布情况。
4. 不稳定的结果
- 画图展示
def plot_clusterer_comparison(c1,c2,X):
c1.fit(X)
c2.fit(X)
plt.figure(figsize=(12,4))
plt.subplot(121)
plot_decision_boundaries(c1,X)
plt.subplot(122)
plot_decision_boundaries(c2,X)
# 创建对象,进行不同(质心初始化的随机数生成)结果对比
c1 = KMeans(n_clusters = 5,init='random',n_init = 1,random_state=11)
c2 = KMeans(n_clusters = 5,init='random',n_init = 1,random_state=19)
plot_clusterer_comparison(c1,c2,X)
-
效果展示
不同的质心初始化的随机数生成次数,得到的结果是不稳定的
5. 评估方法
Inertia指标:每个样本到它最近簇的距离的平方和然后求和
kmeans.inertia_
## 211.5985372581684
X_dist = kmeans.transform(X)
# 找到每个样本到最近的簇的距离
X_dist[np.arange(len(X_dist)),kmeans.labels_]
np.sum(X_dist[np.arange(len(X_dist)),kmeans.labels_]**2)
## 211.59853725816868
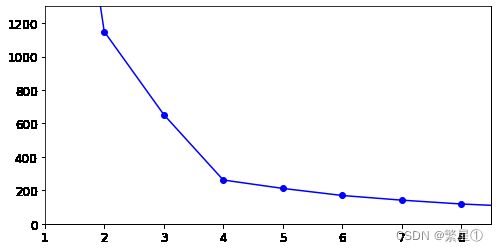
6. 找到最佳簇数
- 选择10次数据进行查找
kmeans_per_k = [KMeans(n_clusters = k).fit(X) for k in range(1,10)]
inertias = [model.inertia_ for model in kmeans_per_k]
- 画图展示
plt.figure(figsize=(8,4))
plt.plot(range(1,10),inertias,'bo-')
plt.axis([1,8.5,0,1300])
plt.show()
7. 轮廓系数
结论:
- s(i)接近1,则说明样本(i)聚类合理;
- s(i)接近-1,则说明样本(i)更应该分类到另外的簇;
- 若s(i) 近似为0,则说明样本(i)在两个簇的边界上。
- 计算所有样本的平均轮廓系数
from sklearn.metrics import silhouette_score
# 计算所有样本的平均轮廓系数
silhouette_score(X,kmeans.labels_)
- 计算所有轮廓系数
silhouette_scores = [silhouette_score(X,model.labels_) for model in kmeans_per_k[1:]]
- 画图展示
plt.figure(figsize=(8,4))
plt.plot(range(2,10),silhouette_scores,'bo-')
# plt.axis([1,8.5,0,1300])
plt.show()
8. Kmeans存在的问题
- 生成原始数据集
X1, y1 = make_blobs(n_samples=1000, centers=((4, -4), (0, 0)), random_state=42)
X1 = X1.dot(np.array([[0.374, 0.95], [0.732, 0.598]]))
X2, y2 = make_blobs(n_samples=250, centers=1, random_state=42)
X2 = X2 + [6, -8]
X = np.r_[X1, X2]
y = np.r_[y1, y2]
plot_data(X)
kmeans_good = KMeans(n_clusters = 3,init = np.array([[-1.5,2.5],[0.5,0],[4,0]]),n_init=1,random_state=42)
kmeans_bad = KMeans(n_clusters = 3,random_state=42)
kmeans_good.fit(X)
kmeans_bad.fit(X)
- 绘制图形
plt.figure(figsize=(10,4))
plt.subplot(121)
plot_decision_boundaries(kmeans_good,X)
# plt.title('Good - inertia = {}'.fotmat(kmeans_good.inertia_))
plt.subplot(122)
plot_decision_boundaries(kmeans_bad,X)
# plt.title('Bad - inertia = {}'.fotmat(kmeans_bad.inertia_))
9. 图像分割实例
- 加载图片
# ladybug.png
from matplotlib.image import imread
image = imread('ladybug.png')
image.shape
- 重塑数据
X = image.reshape(-1,3)
- 进行训练
# 进行训练
kmeans = KMeans(n_clusters = 8,random_state=42).fit(X)
# 八个中心位置
# kmeans.cluster_centers_
- 把原始图像的像素数转化为特定的类别
# 通过标签找到对应的中心点,并且把数据还原成三维数据
segmented_img = kmeans.cluster_centers_[kmeans.labels_].reshape(533, 800, 3)
# 此时的图像当中只有八种不同的像素点
segmented_img
- 把原始数据分为不同的簇,以进行对比实验
segmented_imgs = []
n_colors = (10,8,6,4,2)
for n_clusters in n_colors:
kmeans = KMeans(n_clusters,random_state=42).fit(X)
segmented_img = kmeans.cluster_centers_[kmeans.labels_]
segmented_imgs.append(segmented_img.reshape(image.shape))
- 绘制图形
plt.figure(figsize = (10,5))
plt.subplot(231)
plt.imshow(image)
plt.title('Original image')
for idx,n_clusters in enumerate(n_colors):
plt.subplot(232+idx)
plt.imshow(segmented_imgs[idx])
plt.title('{}colors'.format(n_clusters))
10. 半监督学习
-
加载数据集并对数据集进行拆分
首先,让我们将训练集聚类为50个集群,然后对于每个聚类,让我们找到最靠近质心的图像。 我们将这些图像称为代表性图像:
from sklearn.datasets import load_digits
# 加载并返回数字数据集(分类)
X_digits,y_digits = load_digits(return_X_y = True)
from sklearn.model_selection import train_test_split
# 将数组或矩阵切分为随机训练和测试子集
X_train,X_test,y_train,y_test = train_test_split(X_digits,y_digits,random_state=42)
- 进行训练
- 第一类:直接选择50个样本进行逻辑回归的计算
# 使用逻辑回归进行半监督学习
from sklearn.linear_model import LogisticRegression
n_labeled = 50
log_reg = LogisticRegression(random_state=42)
log_reg.fit(X_train[:n_labeled], y_train[:n_labeled])
log_reg.score(X_test, y_test)
结果:0.8266666666666667
- 第二类:用聚类算法找到距离簇中心最近的具有代表性的点,再进行逻辑回归的计算
# 对训练数据进行聚类
k = 50
kmeans = KMeans(n_clusters=k, random_state=42)
# 得到1347个样本到每个簇的距离
X_digits_dist = kmeans.fit_transform(X_train)
# 找簇中距离质心最近的点的索引
representative_digits_idx = np.argmin(X_digits_dist,axis=0)
# 把索引回传到训练样本中,找到当前的数据
X_representative_digits = X_train[representative_digits_idx]
现在让我们绘制这些代表性图像并手动标记它们:
# 画图并展示
plt.figure(figsize=(8, 2))
for index, X_representative_digit in enumerate(X_representative_digits):
plt.subplot(k // 10, 10, index + 1)
plt.imshow(X_representative_digit.reshape(8, 8), cmap="binary", interpolation="bilinear")
plt.axis('off')
plt.show()
根据以上结果进行手动标记标签:
# 手动打标签
y_representative_digits = np.array([
4, 8, 0, 6, 8, 3, 7, 7, 9, 2,
5, 5, 8, 5, 2, 1, 2, 9, 6, 1,
1, 6, 9, 0, 8, 3, 0, 7, 4, 1,
6, 5, 2, 4, 1, 8, 6, 3, 9, 2,
4, 2, 9, 4, 7, 6, 2, 3, 1, 1])
现在我们有一个只有50个标记实例的数据集,它们中的每一个都是其集群的代表性图像,而不是完全随机的实例,然后去训练:
log_reg = LogisticRegression(random_state=42)
log_reg.fit(X_representative_digits, y_representative_digits)
log_reg.score(X_test, y_test)
结果:0.92
- 第三类:将标签传播到同一群集中的所有其他实例,然后进行逻辑回归的计算
# 先做一个空的标签
y_train_propagated = np.empty(len(X_train), dtype=np.int32)
# 遍历打标签
for i in range(k):
y_train_propagated[kmeans.labels_==i] = y_representative_digits[i]
log_reg = LogisticRegression(random_state=42)
log_reg.fit(X_train, y_train_propagated)
log_reg.score(X_test, y_test)
结果:0.9288888888888889
- 第四类:选择前20个来进行逻辑回归的计算
## 核心:簇 -> 索引 -> 样本
percentile_closest = 20
X_cluster_dist = X_digits_dist[np.arange(len(X_train)), kmeans.labels_]
for i in range(k):
in_cluster = (kmeans.labels_ == i)
# 选择属于当前簇的所有样本
cluster_dist = X_cluster_dist[in_cluster]
# 排序找到前20个
cutoff_distance = np.percentile(cluster_dist, percentile_closest)
# False True结果
above_cutoff = (X_cluster_dist > cutoff_distance)
X_cluster_dist[in_cluster & above_cutoff] = -1
# 找到索引
partially_propagated = (X_cluster_dist != -1)
# 回传到样本
X_train_partially_propagated = X_train[partially_propagated]
y_train_partially_propagated = y_train_propagated[partially_propagated]
# 进行训练
log_reg = LogisticRegression(random_state=42)
log_reg.fit(X_train_partially_propagated, y_train_partially_propagated)
log_reg.score(X_test, y_test)
结果:0.9444444444444444
11. DBSCAN算法
- 准备数据集
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=1000, noise=0.05, random_state=42)
- 导包并进行对比实验
from sklearn.cluster import DBSCAN
# eps:半径值,min_samples:一个点被视为核心点的邻域内的样本数
# 进行对比实验
dbscan = DBSCAN(eps = 0.05,min_samples=5)
dbscan.fit(X)
dbscan2 = DBSCAN(eps = 0.2,min_samples=5)
dbscan2.fit(X)
- 绘制图形
def plot_dbscan(dbscan, X, size, show_xlabels=True, show_ylabels=True):
core_mask = np.zeros_like(dbscan.labels_, dtype=bool)
core_mask[dbscan.core_sample_indices_] = True
anomalies_mask = dbscan.labels_ == -1
non_core_mask = ~(core_mask | anomalies_mask)
cores = dbscan.components_
anomalies = X[anomalies_mask]
non_cores = X[non_core_mask]
plt.scatter(cores[:, 0], cores[:, 1],
c=dbscan.labels_[core_mask], marker='o', s=size, cmap="Paired")
plt.scatter(cores[:, 0], cores[:, 1], marker='*', s=20, c=dbscan.labels_[core_mask])
plt.scatter(anomalies[:, 0], anomalies[:, 1],
c="r", marker="x", s=100)
plt.scatter(non_cores[:, 0], non_cores[:, 1], c=dbscan.labels_[non_core_mask], marker=".")
if show_xlabels:
plt.xlabel("$x_1$", fontsize=14)
else:
plt.tick_params(labelbottom='off')
if show_ylabels:
plt.ylabel("$x_2$", fontsize=14, rotation=0)
else:
plt.tick_params(labelleft='off')
plt.title("eps={:.2f}, min_samples={}".format(dbscan.eps, dbscan.min_samples), fontsize=14)
plt.figure(figsize=(18, 4))
plt.subplot(131)
plt.plot(X[:,0],X[:,1],'b.')
plt.xlabel("$x_1$", fontsize=14)
plt.ylabel("$x_2$", fontsize=14,rotation=0)
plt.title("Original image")
plt.subplot(132)
plot_dbscan(dbscan, X, size=100)
plt.subplot(133)
plot_dbscan(dbscan2, X, size=600, show_ylabels=True)
plt.show()