使用MD.ai标注医学图像及covid-19数据集标注
本文档涵盖注册、推荐的浏览器设置、打开分配的案例和故障排除。了解使用 MD.ai 启动和运行所有深度学习项目的基础知识。
1、登录
打开浏览器:https://www.md.ai并单击Create Account按钮。
登录中
如果您收到项目的电子邮件邀请,只需单击电子邮件中的链接即可访问。确保使用邀请中的电子邮件进行登录。
例如,如果您收到了对您.edu地址的邀请,并且您尝试使用您的 gmail 帐户登录,则不会授予访问权限。使用正确的电子邮件登录将成功打开项目。
浏览器
MD.ai 注释器界面是针对最新的 Google Chrome 浏览器优化的 HTML5 Web 应用程序。我们强烈建议在笔记本电脑/台式机上使用 Chrome。
Annotator 已在 MacOS、ChromeOS 和 Ubuntu Linux 上进行了最广泛的测试,我们建议至少 8GB 的 RAM。
不支持使用移动设备进行注释,但对于全局级别的注释可能已足够。Mozilla Firefox 尚未经过测试,但应该可以正常工作。
2、项目界面
顶部工具栏有以下选项
项目专栏
标签栏
导航栏
更新栏
顶行按钮
查看器设置
使用多种配置在 1-6 个查看器中查看系列或图像。
您还可以从两个系列同步到六个系列以进行同步滚动。具有不同方向的系列将显示非活动系列的
参考线。
参考线
全屏模式
在全屏视图中打开注释器。
将注释/标签添加到活动注释
将注释或 radlex 标签添加到当前活动的注释。如果没有激活的注释,此选项将不可用。
将活动注释复制到上一个/下一个图像
将活动注释复制到上一个/下一个图像。然后,您可以单独编辑、重绘或删除注释。如果没有激活的注释,此选项将不可用。
撤销重做
切换最近的更改。
清除活动层
删除形状。
画笔工具
画笔工具包括画笔宽度和形状以及用于修正的橡皮擦。.
擦除
将擦除自由形状或多边形上的各个点。
网格
显示网格标记。您可以选择正方形或六边形网格。使用网格大小图标或使用键盘快捷键调整网格大小。
统治者
添加尺子以毫米为单位进行测量。
角度
添加角度以度数为单位测量角度。
椭圆
添加椭圆 ROI 工具。
像素探头
显示单个点的像素值。
飞涨
放大或缩小图像。
逆时针/顺时针旋转
将系列中的所有图像逆时针或顺时针旋转 90 度。
水平/垂直翻转
沿水平/垂直轴依次翻转所有图像。
居中并重置视口
重置对图像所做的所有更改,包括大小、旋转、窗口/级别或过滤器。
反转颜色
反转图像颜色。
窗口/水平
更改窗口和级别设置。还有键盘快捷键可以设置 CT 扫描的窗口和级别。
ROI 窗口/级别
针对图像上感兴趣的区域自动优化窗口和级别设置。
图像过滤器
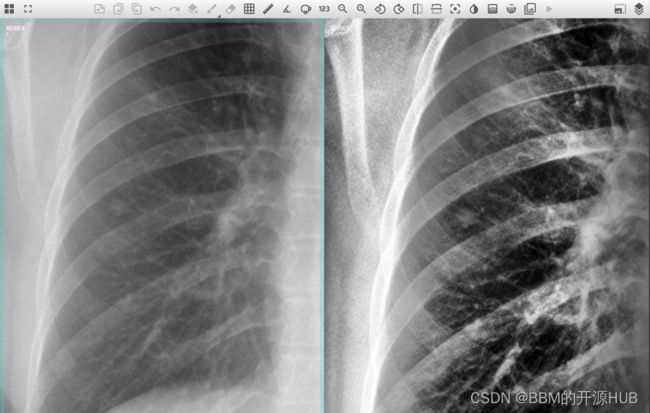
不同的图像过滤器,包括锐化、模糊、浮雕、边缘 (sobel) 和直方图均衡
直方图均衡化 (CLAHE) 前后的示例 CXR,以突出显示右肺中部的细微肺结节。
右肺中部细微肺结节的特写。
电影播放
以电影模式连续显示图像。选择电影速度和方向。
图像渲染设置
在滚动过程中图像被替换为低分辨率版本,并在滚动或电影停止时通过设置返回高分辨率版本Use Rendered Image。
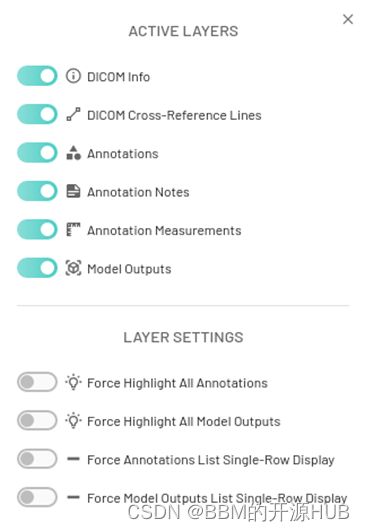
图层控制
隐藏或显示 dicom 信息、交叉参考线、注释、注释、测量和模型输出。还有在单行中显示注释/模型输出的选项。
3、数据集
同时为大家准备了经过md.ai标注的covid-19数据集,感兴趣的小伙伴们可以通过下面的地址下载,因为数据集太大,我把数据集放在了百度网盘,为保证网盘容量需要定期支付百度网盘容量使用费,因此该下载需要收取一定的费用,数据集有三部分组成,分别为1a、1b、1c三个版本,下载地址如下:
MIDRC-RICORD-1a-数据集文档类资源-CSDN下载
数据摘要
1. 120 次胸部 CT 检查(仅限轴向系列,任何协议)。
2. 注释包括
a) 受影响区域的详细分割;
b) 图像级标签(感染性不透明度、感染性 TIB/微结节、感染性腔、非感染性结节/肿块、肺不张、其他非感染性不透明度)
c) 检查级诊断标签(典型、不确定、非典型、肺炎阴性、晕征、反转晕征、无实质混浊的网状模式、病灶周围血管扩大、支气管壁增厚、支气管扩张、胸膜下曲线线、积液、胸膜增厚、气胸、心包积液、淋巴结病、肺栓塞、正常肺、感染性肺病、肺气肿、肿瘤性肺病、非感染性炎症性肺病、非感染性间质性肺病、纤维化肺病、其他肺病)
d) 检查级程序标签(有 IV 造影剂,没有 IV 造影剂,QA- 运动/呼吸不足,QA- 吸气不足,QA- 低分辨率不足,QA- 肺不完整,QA- 身体部位/模态不足,气管导管、中央静脉/动脉导管、鼻胃管、胸骨切开线、起搏器、其他支持装置)。
3. 支持临床变量:MRN*、年龄、研究日期*、检查描述、性别、研究 UID*、图像计数、模态、测试结果、样本来源(* 假名值)。
MIDRC-RICORD-1b-数据集文档类资源-CSDN下载
本文档涵盖注册、推荐的浏览器设置、打开分配的案例和故障排除。了解使用 MD.ai 启动和运行所有深度学习项目的基础知识。
MIDRC-RICORD-1c-数据集文档类资源-CSDN下载
数据摘要
(1)、来自 361 名患者的 998 次胸部 X 光检查。
(2)、带标签的注释:
a、分类
典型外观
多灶性双侧、外周混浊和/或圆形混浊
下肺为主的分布(必需特征 - 必须存在前两种混浊模式中的一种或两种)
不确定
的外观 没有典型的发现和单侧,中央或上肺主要分布的气腔疾病
非典型表现
气胸或胸腔积液、肺水肿、肺叶实变、孤立性肺结节或肿块、弥漫性微小结节、空洞
肺炎阴性
无肺部混浊
b、空域疾病分级
肺在正面胸部 X 光片上被分成每肺 3 个区域(总共 6 个区域)。上部区域从顶点延伸到上肺门。中区跨越肺门上缘和下缘。下部区域从肺门下缘延伸至肋膈沟。
*轻度- 如果肺炎不是阴性,则需要1-2 个肺区混浊
*中度- 如果肺炎不是阴性,则需要3-4 个肺区混浊
*严重- 如果肺炎不是阴性,则需要>4 个肺区混浊
(3)、支持临床变量:MRN*、年龄、研究日期*、检查描述、性别、研究 UID*、图像计数、模态、测试结果、样本来源(* 假名值)。
4、数据转换
(1)将批注 MD.ai 转换为蒙版
用于从一行注释数据加载单个掩码实例的函数。这会将一个框,自由形式,多边形等转换为与相应图像大小相关的二进制蒙版。
def load_mask_instance(row):
"""Load instance masks for the given annotation row. Masks can be different types,
mask is a binary true/false map of the same size as the image.
"""
mask = np.zeros((row.height, row.width), dtype=np.uint8)
annotation_mode = row.annotationMode
# print(annotation_mode)
if annotation_mode == "bbox":
# Bounding Box
x = int(row["data"]["x"])
y = int(row["data"]["y"])
w = int(row["data"]["width"])
h = int(row["data"]["height"])
mask_instance = mask[:,:].copy()
cv2.rectangle(mask_instance, (x, y), (x + w, y + h), 255, -1)
mask[:,:] = mask_instance
# FreeForm or Polygon
elif annotation_mode == "freeform" or annotation_mode == "polygon":
vertices = np.array(row["data"]["vertices"])
vertices = vertices.reshape((-1, 2))
mask_instance = mask[:,:].copy()
cv2.fillPoly(mask_instance, np.int32([vertices]), (255, 255, 255))
mask[:,:] = mask_instance
# Line
elif annotation_mode == "line":
vertices = np.array(row["data"]["vertices"])
vertices = vertices.reshape((-1, 2))
mask_instance = mask[:,:].copy()
cv2.polylines(mask_instance, np.int32([vertices]), False, (255, 255, 255), 12)
mask[:,:] = mask_instance
elif annotation_mode == "location":
# Bounding Box
x = int(row["data"]["x"])
y = int(row["data"]["y"])
mask_instance = mask[:,:].copy()
cv2.circle(mask_instance, (x, y), 7, (255, 255, 255), -1)
mask[:,:] = mask_instance
elif annotation_mode is None:
print("Not a local instance")
return mask.astype(np.bool)(2)将md.ai的json文件转化为dcm的seg文件
import argparse
import distutils.spawn
import os
import pandas as pd
import numpy as np
import mdai
import logging
import pydicom
import tqdm
import itk
import cv2
import copy
import glob
import subprocess
import glob
# logging setup
logging.basicConfig()
# By default the root logger is set to WARNING and all loggers you define
# inherit that value. Here we set the root logger to NOTSET. This logging
# level is automatically inherited by all existing and new sub-loggers
# that do not set a less verbose level.
logging.root.setLevel(logging.NOTSET)
# The following line sets the root logger level as well.
# It's equivalent to both previous statements combined:
logging.basicConfig(level=logging.NOTSET)
logger = logging.getLogger('dcmqi.mdai2dcm')
logger.setLevel(logging.ERROR)
# Helper functions
dcmqi_template = {
"ContentCreatorName": "",
"ClinicalTrialSeriesID": "Session1",
"ClinicalTrialTimePointID": "1",
"SeriesDescription": "Segmentation",
"SeriesNumber": "300",
"InstanceNumber": "1",
"BodyPartExamined": "CHEST",
"segmentAttributes": [
],
"ContentLabel": "SEGMENTATION",
"ContentDescription": "Image segmentation",
"ClinicalTrialCoordinatingCenterName": "dcmqi"
}
def makeHash(text, length=6):
from base64 import b64encode
from hashlib import sha1
return b64encode(sha1(str.encode(text)).digest()).decode('ascii')[:length]
segment_template = {
"labelID": "",
"SegmentDescription": "",
"SegmentAlgorithmType": "MANUAL",
"SegmentedPropertyCategoryCodeSequence": {
"CodeValue": "49755003",
"CodingSchemeDesignator": "SCT",
"CodeMeaning": "Morphologically Abnormal Structure"
},
"SegmentedPropertyTypeCodeSequence": {
"CodeValue": "",
"CodingSchemeDesignator": "99RICORD",
"CodeMeaning": ""
},
"recommendedDisplayRGBValue": [
177,
122,
101
]
}
def convertToNIfTI(series_dir, reconstruction_dir):
# if nii file exist return it
logger.debug("Saving reconstruction to ", reconstruction_dir)
reconstruction = reconstruction_dir+'/ct_image.nii'
if os.path.exists(reconstruction):
return reconstruction
# else create one with dcm2niix
cmd = ['dcm2niix', '-o', reconstruction_dir, '-f', 'ct_image', series_dir]
subprocess.run(cmd, stderr=subprocess.DEVNULL, stdout=subprocess.DEVNULL)
return reconstruction
def convertToSEG2(input_dicom_dir, seg_dir):
logger.debug("Saving DICOM SEG to "+seg_dir)
json_files = glob.glob(seg_dir+"/*.json")
for label_json in json_files:
creator = os.path.split(label_json)[1].split('-')[0]
label_dcm = os.path.join(seg_dir,creator+".dcm")
labels = glob.glob(seg_dir+'/'+creator+'*.nii')
labels.sort()
cmd = ['itkimage2segimage','--inputDICOMDirectory',input_dicom_dir,
'--inputImageList',','.join(labels),"--inputMetadata",
label_json, "--outputDICOM", label_dcm, "--skip"]
logger.debug('Running SEG conversion with:\n'+' '.join(cmd))
result = subprocess.run(cmd, stderr=subprocess.PIPE, stdout=subprocess.PIPE, text=True)
logger.debug(result.stderr)
logger.debug(result.stdout)
def main():
parser = argparse.ArgumentParser(
usage="%(prog)s --inputDICOM --inputJSON --outputDirectory \n\n"
"Warning: This is an experimenta script in development!\n"
"The intent of this helper script is to enable conversion"
"of the MD.ai annotations into appropriate standard DICOM objects.\n")
parser.add_argument(
'--inputDICOM',
dest="inputDICOM",
default="/Users/fedorov/github/mdai2seg/mydata/mdai_rsna_project_G9qOEdR0_images_2020-10-27-080731",
metavar='Input directory with the DICOM images being annotated',
help="Directory with the input DICOM images. It is expected that"
" the content of this directory is organized into the following"
"hierarchy: //.dcm")
#,
#required=True)
parser.add_argument(
'--inputJSON',
dest="inputJSON",
default="/Users/fedorov/github/mdai2seg/mydata/mdai_rsna_project_G9qOEdR0_annotations_labelgroup_all_2020-10-27-080732.json",
metavar='Input MD.ai annotations in the native JSON representation',
help="MD.ai nnotations stored in MD.ai format.")
#,
#required=True)
parser.add_argument(
'--outputDirectory',
dest="outputDirectory",
default="/Users/fedorov/Downloads/test_seg2",
metavar='Output directory to store the resulting DICOM files',
help="Directory to store resulting converted DICOM objects.")
#,
#required=True)
parser.add_argument(
'--includedLabelGroups',
dest="includedLabelGroups",
default="\"Team Emily\",\"Team Scott\"",
metavar='Label groups to consider in conversion',
help="Only the specified label groups will be considered.")
#,
#required=True)
args = parser.parse_args()
if not os.path.exists(args.outputDirectory):
os.mkdir(args.outputDirectory)
results = mdai.common_utils.json_to_dataframe(args.inputJSON)
annotations_df = results['annotations']
logger.info(f"{annotations_df.shape[0]} annotations loaded")
all_series_uids = annotations_df['SeriesInstanceUID'].unique()
# TODO: Why there are NaNs?
all_series_uids = all_series_uids[~pd.isnull(all_series_uids)]
progress_bar = tqdm.tqdm(total=len(all_series_uids))
for this_series_uid in all_series_uids:
#['1.2.826.0.1.3680043.10.474.419639.300423266679936330916265249312']: # ['1.2.826.0.1.3680043.10.474.440808.1993']: #all_series_uids: # ['1.2.826.0.1.3680043.10.474.2969551981555819856670502082591727602']:
logger.debug(f"Processing {this_series_uid}")
# filter our irrelevant rows
this_series_annotations = annotations_df[annotations_df["SeriesInstanceUID"] == this_series_uid]
this_series_annotations = this_series_annotations[this_series_annotations['annotationMode'] == 'freeform']
this_series_annotations = this_series_annotations[this_series_annotations['groupName'].isin(['Team Emily', 'Team Scott'])]
if this_series_annotations.shape[0] == 0:
continue
logger.debug('Creators of freeform annotations for series '+this_series_uid+" are "+str(this_series_annotations['createdById'].unique()))
one_row = this_series_annotations.iloc[0]
series_uid = one_row["SeriesInstanceUID"]
study_path = os.path.join(args.inputDICOM,one_row["StudyInstanceUID"])
segmentations_path = os.path.join(args.outputDirectory,series_uid+"_SEG")
reconstruction_path = os.path.join(args.outputDirectory,series_uid+"_Reconstruction")
if not os.path.exists(segmentations_path):
os.mkdir(segmentations_path)
if not os.path.exists(reconstruction_path):
os.mkdir(reconstruction_path)
series_path = os.path.join(study_path,one_row["SeriesInstanceUID"])
path_to_instance = os.path.join(series_path,one_row["SOPInstanceUID"]+".dcm")
instance_dcm = pydicom.dcmread(path_to_instance)
ct_image_file_name = convertToNIfTI(series_path, reconstruction_path) # run dcm2niix to do this conversion
PixelType = itk.ctype("signed short")
image_volume = itk.imread(ct_image_file_name, PixelType)
image_volume_array = itk.array_view_from_image(image_volume)
label_images_per_creator = {}
label_json_per_creator = {}
creators = this_series_annotations['createdById'].unique()
for creator in creators:
label_images_per_creator[creator] = []
label_json_per_creator[creator] = copy.deepcopy(dcmqi_template)
label_json_per_creator[creator]['ContentCreatorName'] = creator
label_json_per_creator[creator]['segmentAttributes'] = []
for index, row in this_series_annotations.iterrows():
if row['annotationMode'] != 'freeform':
progress_bar.update(1)
continue
if row['data'] == None or row['data']['vertices'] == None:
progress_bar.update(1)
continue
instance_uid = row['SOPInstanceUID']
creator = row['createdById']
path_to_instance = os.path.join(args.inputDICOM,row["StudyInstanceUID"],row["SeriesInstanceUID"],row["SOPInstanceUID"]+".dcm")
instance_dcm = pydicom.dcmread(path_to_instance)
slice_origin_physical_point = instance_dcm.ImagePositionPatient
slice_origin_index = image_volume.TransformPhysicalPointToIndex(slice_origin_physical_point)
z = slice_origin_index[2]
# make a separate image from single slice
extractor = itk.ExtractImageFilter.New(image_volume)
region = image_volume.GetLargestPossibleRegion()
size = region.GetSize()
size[2] = 1
slice_origin_index[0] = 0
slice_origin_index[1] = 0
region.SetIndex(slice_origin_index)
region.SetSize(size)
extractor.SetExtractionRegion(region)
extractor.SetInput(image_volume)
extractor.Update()
label_slice_image = extractor.GetOutput()
label_slice_array = itk.array_from_image(label_slice_image)
label_slice_array.fill(0)
vertices = np.array(row['data']['vertices']).reshape((-1, 2))
slice_label_mask = label_slice_array[0,:,:]
cv2.fillPoly(slice_label_mask, np.int32([vertices]), 1)
label_slice_array[0,:,:]=np.flipud(slice_label_mask)
new_image = itk.image_from_array(label_slice_array)
new_image.SetDirection(label_slice_image.GetDirection())
new_image.SetOrigin(label_slice_image.TransformIndexToPhysicalPoint(slice_origin_index))
new_image.SetSpacing(label_slice_image.GetSpacing())
#logger.debug("added annotation for creator "+creator)
label_images_per_creator[creator].append(new_image)
this_segment_template = copy.deepcopy(segment_template)
this_segment_template['labelID'] = 1
this_segment_template['SegmentDescription'] = row['labelName']
this_segment_template['recommendedDisplayRGBValue'] = [int(row['color'][1:3],16), int(row['color'][3:5], 16), int(row['color'][5:7],16)]
this_segment_template['SegmentedPropertyTypeCodeSequence']['CodeValue'] = makeHash(row['labelName'])
this_segment_template['SegmentedPropertyTypeCodeSequence']['CodeMeaning'] = row['labelName']
label_json_per_creator[creator]['segmentAttributes'].append([this_segment_template])
for creator in creators:
logger.debug('Creator '+creator+' has '+str(len(label_images_per_creator[creator]))+' segmentations and '+str(len(label_json_per_creator[creator]['segmentAttributes']))+' seg attrs')
logger.debug('Processing labels for '+creator)
for idx, label_itk_image in enumerate(label_images_per_creator[creator]):
output_file_name = creator+"-"+('%03d' % idx)
itk.imwrite(label_itk_image, os.path.join(segmentations_path,output_file_name+".nii"))
import json
output_json_name = creator+"-metadata.json"
with open(os.path.join(segmentations_path,output_json_name), "w") as json_file:
json_file.write(json.dumps(label_json_per_creator[creator], indent=2))
convertToSEG2(series_path, segmentations_path)
if len(glob.glob(segmentations_path+"/*dcm")) == 0:
logger.error(f"SEG conversion failed for {segmentations_path}!")
progress_bar.update(1)
logger.debug("Done")
return
if __name__ == "__main__":
exeFound = {}
for exe in ['dcm2niix', 'itkimage2segimage']:
if distutils.spawn.find_executable(exe) is None:
exeFound[exe] = False
else:
exeFound[exe] = True
if not(exeFound['itkimage2segimage']) or not (exeFound['dcm2niix']):
logger.error(
"Dependency converter(s) not found in the path.")
logger.error(
"dcmqi (https://github.com/qiicr/dcmqi) and dcm2niix (https://github.com/rordenlab/dcm2niix/releases)")
logger.error(
"need to be installed and available in the PATH for using this converter script.")
sys.exit()
main() 从 MD.ai JSON 转换为 DICOM-SEG 的代码:https ://github.com/QIICR/dcmqi/blob/add-mdai-converter/util/mdai2dcm.py