利用python从蛋白文件中提取特定类型的原子
python从蛋白文件中提取特定类型的原子
文章目录
- python从蛋白文件中提取特定类型的原子
- 前言
- 一、代码
- 二、使用方式
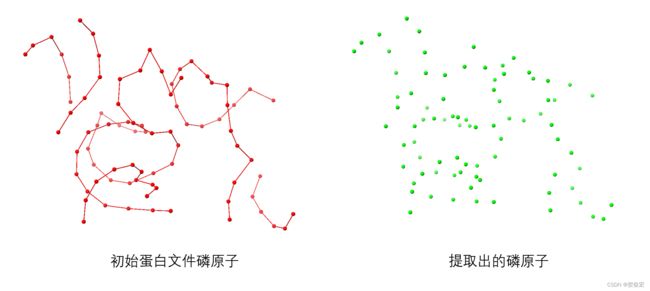
- 三. 结果展示
前言
因为项目的需要,需要从蛋白文件中提取特定的某一中元素(本代码以P原子为例),在此记录一下代码
一、代码
import os, argparse
import numpy as np
PDB_CHAIN_IDS = 'ABCDEFGHIJKLMNOPQRSTUVWXYZabcdefghijklmnopqrstuvwxyz0123456789'
def read_atom_line(line):
"""
get infomation the line of pdb file
Args:
ciffile: the line of pdb file (str)
"""
if len(line) > 78:
line = line.strip()
assert line.startswith('ATOM')
atom_idx = line[6:11].strip()
atom_name = line[12:16].strip()
restype = line[17:20].strip()
chain = line[21]
res_index = line[22:26].strip()
x, y, z = float(line[30:38].strip()), float(line[38:46].strip()), float(line[46:54].strip())
occ = float(line[54:60].strip())
temp = float(line[60:66].strip())
return atom_idx, atom_name, restype, chain, res_index, x, y, z, occ, temp
def read_pdb(pdbfile, aatype):
"""
extract infomation of a specific atomic type from the pdb file
Args:
ciffile: the path of cif file. (str)
aatype: the type of atom. (str)
"""
x_list, y_list, z_list, occ_lis, temp_list, restype_list = [], [], [], [], [], []
for line in open(pdbfile):
if line.startswith('ATOM'):
atom_idx, atom_name, restype, chain, res_index, x, y, z, occ, temp = read_atom_line(line)
if atom_name == aatype:
atom_coord_list.append([x, y, z])
occ_list.append(occ)
temp_list.append(temp)
restype_list.append(restype)
x_list = np.array(x_list)
y_list = np.array(y_list)
z_list = np.array(z_list)
occ_list = np.array(occ_list)
temp_list = np.array(temp_list)
restype_list = np.array(restype_list)
return x_list, y_list, z_list, restype_list, occ_list, temp_list
def read_cif(ciffile, aatype):
"""
extract infomation of a specific atomic type from the cif file
Args:
ciffile: the path of cif file. (str)
aatype: the type of atom. (str)
"""
mmcif_dict = Bio.PDB.MMCIF2Dict.MMCIF2Dict(ciffile)
cond = (np.array(mmcif_dict['_atom_site.label_atom_id']) == 'P')
x_list = np.array(mmcif_dict['_atom_site.Cartn_x'])[cond]
y_list = np.array(mmcif_dict['_atom_site.Cartn_y'])[cond]
z_list = np.array(mmcif_dict['_atom_site.Cartn_z'])[cond]
occ_list = np.array(mmcif_dict['_atom_site.occupancy'])[cond]
temp_list = np.array(mmcif_dict['_atom_site.B_iso_or_equiv'])[cond]
restype_list = np.array(mmcif_dict['_atom_site.label_comp_id'])[cond]
return x_list, y_list, z_list, restype_list, occ_list, temp_list
def extract_atom_to_pdb(input_protein, output_pdb, aatype='P'):
"""
extract infomation of a specific atomic type from input file and write to pdb file
Args:
input_protein: the path of protein file. (str)
aatype: the type of atom. (str)
"""
out_f = open(output_pdb, 'w')
res_idx = 1
ch_idx = 0
## load info of protein file
assert (os.path.splitext(file)[-1] in ['.cif', '.pdb']), 'input protein file type should be pdb or cif'
if output_pdb.endswith('cif'):
x_list, y_list, z_list, restype_list, occ_list, temp_list = read_cif(input_protein, aatype)
elif output_pdb.endswith('pdb'):
x_list, y_list, z_list, restype_list, occ_list, temp_list = read_pdb(input_protein, aatype)
## write to pdb
protein_info = zip(x_list, y_list, z_list, restype_list, occ_list, temp_list)
for atom_idx, (x, y, z, restype, occ, temp) in enumerate(protein_info):
if res_idx == 9999:
res_idx = 1
ch_idx += 1
if output_pdb:
line = atom_line(atom_idx, aatype, restype,
PDB_CHAIN_IDS[ch_idx], res_idx,
float(x), float(y), float(z),
float(occ), float(temp))
print(line, file=out_f)
print('TER', file=out_f)
res_idx += 1
if __name__ == "__main__":
parser = argparse.ArgumentParser(description='extract specific atomic type of the protein file')
parser.add_argument('aatype', type=str, help="the atom label")
parser.add_argument('input_protei', type=str, help="Input PDB file with CA atoms")
parser.add_argument('output_pdb', type=str, help="Output traced PDB file")
args = parser.parse_args()
aatype = args.aatype
output_pdb = args.output_pdb
input_p_pdb = args.input_p_pdb
extract_atom_to_pdb(input_protein, output_pdb, aatype)
二、使用方式
python xxx.py 蛋白文件 输出文件 原子类型
示例:
python extract_atom.py 7lya.cif 7lya_P.pdb P