MoLeR 分子生成模型,可以指定包含骨架scaffold生成
参考:
https://github.com/microsoft/molecule-generation
https://arxiv.org/pdf/2103.03864.pdf
安装:
注意需要安装tf依赖
pip install molecule-generation
1、使用代码
权重下载,百度网盘 https://pan.baidu.com/s/1lkiWK9-d5MvNyzqRrusGXA?pwd=4hij;保存到example_model_directory文件下
from molecule_generation import load_model_from_directory
model_dir = "./example_model_directory"
example_smiles = ["c1ccccc1", "CNC=O"]
with load_model_from_directory(model_dir) as model:
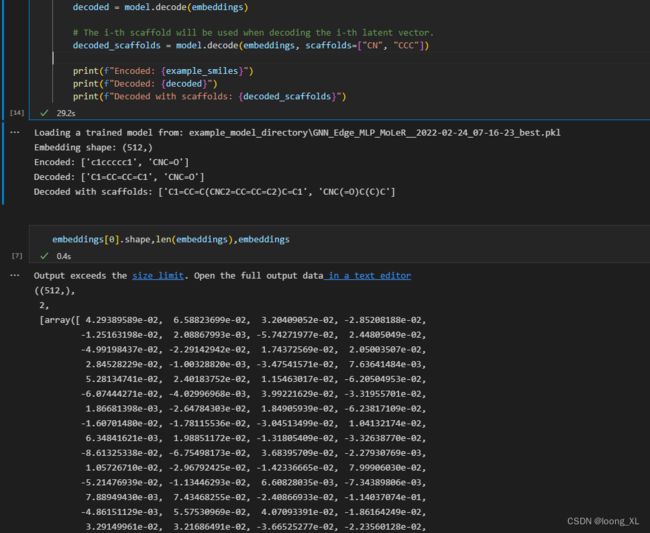
embeddings = model.encode(example_smiles)
print(f"Embedding shape: {embeddings[0].shape}")
# Decode without a scaffold constraint.
decoded = model.decode(embeddings)
# The i-th scaffold will be used when decoding the i-th latent vector.
decoded_scaffolds = model.decode(embeddings, scaffolds=["CN", "CCC"])
print(f"Encoded: {example_smiles}")
print(f"Decoded: {decoded}")
print(f"Decoded with scaffolds: {decoded_scaffolds}")
没指定骨架生成
with load_model_from_directory(model_dir) as model:
embedding2 = model.encode(["O=C1[C@@H](CC)N(CC2=CC(C)=CS2)C3=C(N1C)C=NC(NC4=C(OCC)C=C(C5CCN(C)CC5)C=C4)=N3"])
# The i-th scaffold will be used when decoding the i-th latent vector.
decoded2 = model.decode(embedding2, scaffolds=["N1C(=O)c2ccc(Br)cc2C1=O"])
print(decoded2)
指定骨架生成
with load_model_from_directory(model_dir) as model:
embedding2 = model.encode(["O=C1[C@@H](CC)N(CC2=CC(C)=CS2)C3=C(N1C)C=NC(NC4=C(OCC)C=C(C5CCN(C)CC5)C=C4)=N3"])
# The i-th scaffold will be used when decoding the i-th latent vector.
decoded2 = model.decode(embedding2, scaffolds=["N1C(=O)c2ccc(Br)cc2C1=O"])
print(decoded2)
生成多个相似分子,latent采样附近来生成
1)指定骨架
import numpy as np
from molecule_generation import VaeWrapper
model_dir = "./example_model_directory"
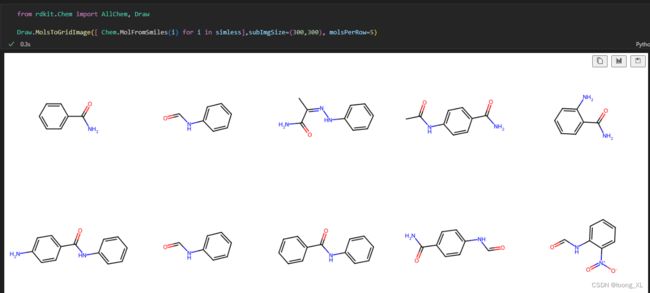
scaffold = "C1=CC=CC=C1"
init_mol = "O=CNC1=CC=CC=C1"
simless =[]
with VaeWrapper(model_dir) as model:
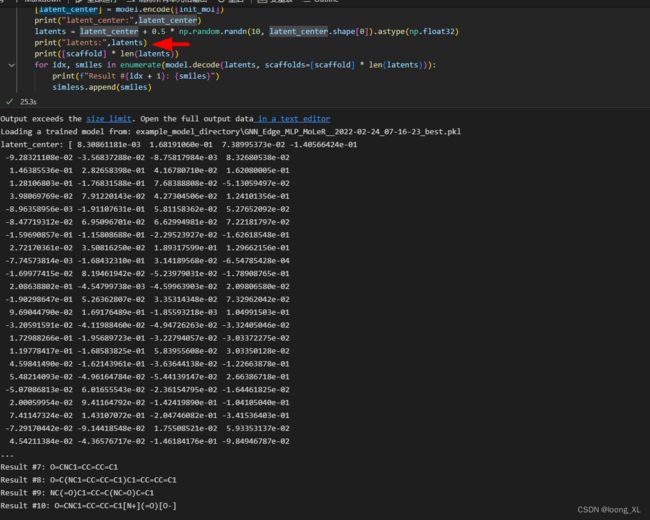
[latent_center] = model.encode([init_mol])
latents = latent_center + 0.5 * np.random.randn(10, latent_center.shape[0]).astype(np.float32) ### 一个latent_center中心,随机latent采样附近10个来生成
# print("latents:",latents)
for idx, smiles in enumerate(model.decode(latents, scaffolds=[scaffold] * len(latents))):
print(f"Result #{idx + 1}: {smiles}")
simless.append(smiles)
from rdkit.Chem import AllChem, Draw
Draw.MolsToGridImage([ Chem.MolFromSmiles(i) for i in simless],subImgSize=(300,300), molsPerRow=5)
import numpy as np
from molecule_generation import VaeWrapper
model_dir = "./example_model_directory"
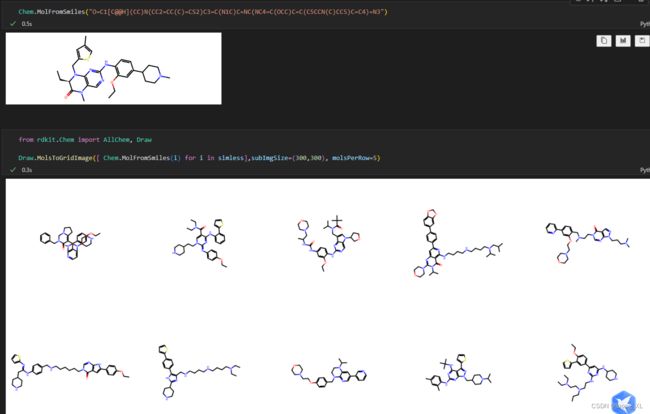
init_mol = "O=C1[C@@H](CC)N(CC2=CC(C)=CS2)C3=C(N1C)C=NC(NC4=C(OCC)C=C(C5CCN(C)CC5)C=C4)=N3"
simless =[]
with VaeWrapper(model_dir) as model:
[latent_center] = model.encode([init_mol])
print("latent_center:",latent_center)
latents = latent_center + 0.5 * np.random.randn(10, latent_center.shape[0]).astype(np.float32)
print("latents:",latents)
print([scaffold] * len(latents))
for idx, smiles in enumerate(model.decode(latents)):
print(f"Result #{idx + 1}: {smiles}")
simless.append(smiles)