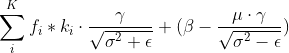RepVGG中
结构重参数化分为两步,第一步是将卷积和BN进行融合以及将只有BN的分支转换成一个Conv2d算子。第二步是将每个分支上的3X3卷积层融合成一个卷积层。
1.1融合Conv和BN
BN中包含四个参数均值![]() 、方差
、方差![]() 、尺度
、尺度![]() 、偏执
、偏执![]() ,对于第i层特征的BN计算公式为:
,对于第i层特征的BN计算公式为:
卷积的形式,这里借用https://blog.csdn.net/qq_37541097/article/details/125692507博主中的图片演示![]()
卷积之后BN的形式是对应位置卷积后减去对应层的均值,然后除以标准差,乘以对应尺度加上偏执,公式大概如:
其中![]() pad后的特征图,
pad后的特征图,![]() 为对应的卷积核,可以将上述公式分解成
为对应的卷积核,可以将上述公式分解成
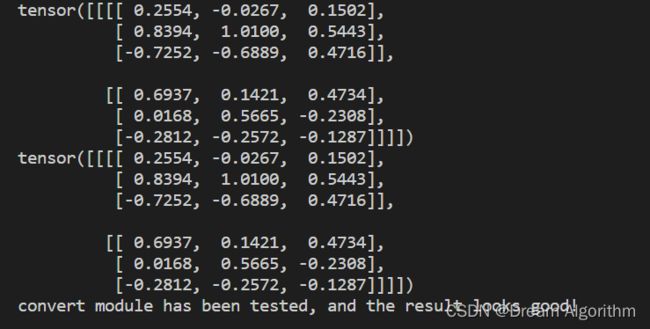
Conv2d+BN融合实验:
from collections import OrderedDict
import numpy as np
import torch
import torch.nn as nn
def main():
torch.random.manual_seed(0)
f1 = torch.randn(1, 2, 3, 3)
module = nn.Sequential(OrderedDict(
conv=nn.Conv2d(in_channels=2, out_channels=2, kernel_size=3, stride=1, padding=1, bias=False),
bn=nn.BatchNorm2d(num_features=2)
))
module.eval()
with torch.no_grad():
output1 = module(f1)
print(output1)
# fuse conv + bn
kernel = module.conv.weight
running_mean = module.bn.running_mean
running_var = module.bn.running_var
gamma = module.bn.weight
beta = module.bn.bias
eps = module.bn.eps
std = (running_var + eps).sqrt()
t = (gamma / std).reshape(-1, 1, 1, 1) # [ch] -> [ch, 1, 1, 1]
kernel = kernel * t
bias = beta - running_mean * gamma / std
fused_conv = nn.Conv2d(in_channels=2, out_channels=2, kernel_size=3, stride=1, padding=1, bias=True)
fused_conv.load_state_dict(OrderedDict(weight=kernel, bias=bias))
with torch.no_grad():
output2 = fused_conv(f1)
print(output2)
np.testing.assert_allclose(output1.numpy(), output2.numpy(), rtol=1e-03, atol=1e-05)
print("convert module has been tested, and the result looks good!")
if __name__ == '__main__':
main()
实验结果:
1.2 将1x1卷积转化成3x3卷积
只需要将原来的权重周围补一圈零,变成和3x3卷积一样的形状,此时padding要从0变为1。
1.3 将BN转化为3x3卷积
对于只有BN层的分支,要构造和3x3卷积一样的形式,才能用上述的形式,这种卷积层是做的恒等映射,输入和输出特征图不变。 输出多少特征图,就用多少个卷积核,每个卷积核只在对应输入通道,对应卷积和通道有值且全为一。![]()
这样就可以将BN转化为Conv2d,然后就可以利用上面的将Conv和BN融合
2、多分枝融合
融合方式很简单,直接将三个分支的参数直接相加
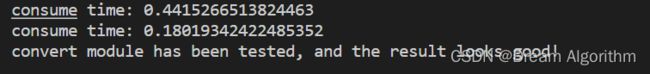
3、结构重参数化实验
import time
import torch.nn as nn
import numpy as np
import torch
def conv_bn(in_channels, out_channels, kernel_size, stride, padding, groups=1):
result = nn.Sequential()
result.add_module('conv', nn.Conv2d(in_channels=in_channels, out_channels=out_channels,
kernel_size=kernel_size, stride=stride, padding=padding,
groups=groups, bias=False))
result.add_module('bn', nn.BatchNorm2d(num_features=out_channels))
return result
class RepVGGBlock(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size=3,
stride=1, padding=1, dilation=1, groups=1, padding_mode='zeros', deploy=False):
super(RepVGGBlock, self).__init__()
self.deploy = deploy
self.groups = groups
self.in_channels = in_channels
self.nonlinearity = nn.ReLU()
if deploy:
self.rbr_reparam = nn.Conv2d(in_channels=in_channels, out_channels=out_channels,
kernel_size=kernel_size, stride=stride,
padding=padding, dilation=dilation, groups=groups,
bias=True, padding_mode=padding_mode)
else:
self.rbr_identity = nn.BatchNorm2d(num_features=in_channels) \
if out_channels == in_channels and stride == 1 else None
self.rbr_dense = conv_bn(in_channels=in_channels, out_channels=out_channels, kernel_size=kernel_size,
stride=stride, padding=padding, groups=groups)
self.rbr_1x1 = conv_bn(in_channels=in_channels, out_channels=out_channels, kernel_size=1,
stride=stride, padding=0, groups=groups)
def forward(self, inputs):
if hasattr(self, 'rbr_reparam'):
return self.nonlinearity(self.rbr_reparam(inputs))
if self.rbr_identity is None:
id_out = 0
else:
id_out = self.rbr_identity(inputs)
return self.nonlinearity(self.rbr_dense(inputs) + self.rbr_1x1(inputs) + id_out)
def get_equivalent_kernel_bias(self):
kernel3x3, bias3x3 = self._fuse_bn_tensor(self.rbr_dense)
kernel1x1, bias1x1 = self._fuse_bn_tensor(self.rbr_1x1)
kernelid, biasid = self._fuse_bn_tensor(self.rbr_identity)
return kernel3x3 + self._pad_1x1_to_3x3_tensor(kernel1x1) + kernelid, bias3x3 + bias1x1 + biasid
def _pad_1x1_to_3x3_tensor(self, kernel1x1):
if kernel1x1 is None:
return 0
else:
return torch.nn.functional.pad(kernel1x1, [1, 1, 1, 1])
def _fuse_bn_tensor(self, branch):
if branch is None:
return 0, 0
if isinstance(branch, nn.Sequential):
kernel = branch.conv.weight
running_mean = branch.bn.running_mean
running_var = branch.bn.running_var
gamma = branch.bn.weight
beta = branch.bn.bias
eps = branch.bn.eps
else:
assert isinstance(branch, nn.BatchNorm2d)
if not hasattr(self, 'id_tensor'):
input_dim = self.in_channels // self.groups
kernel_value = np.zeros((self.in_channels, input_dim, 3, 3), dtype=np.float32)
for i in range(self.in_channels):
kernel_value[i, i % input_dim, 1, 1] = 1
self.id_tensor = torch.from_numpy(kernel_value).to(branch.weight.device)
kernel = self.id_tensor
running_mean = branch.running_mean
running_var = branch.running_var
gamma = branch.weight
beta = branch.bias
eps = branch.eps
std = (running_var + eps).sqrt()
t = (gamma / std).reshape(-1, 1, 1, 1)
return kernel * t, beta - running_mean * gamma / std
def switch_to_deploy(self):
if hasattr(self, 'rbr_reparam'):
return
kernel, bias = self.get_equivalent_kernel_bias()
self.rbr_reparam = nn.Conv2d(in_channels=self.rbr_dense.conv.in_channels,
out_channels=self.rbr_dense.conv.out_channels,
kernel_size=self.rbr_dense.conv.kernel_size, stride=self.rbr_dense.conv.stride,
padding=self.rbr_dense.conv.padding, dilation=self.rbr_dense.conv.dilation,
groups=self.rbr_dense.conv.groups, bias=True)
self.rbr_reparam.weight.data = kernel
self.rbr_reparam.bias.data = bias
for para in self.parameters():
para.detach_()
self.__delattr__('rbr_dense')
self.__delattr__('rbr_1x1')
if hasattr(self, 'rbr_identity'):
self.__delattr__('rbr_identity')
if hasattr(self, 'id_tensor'):
self.__delattr__('id_tensor')
self.deploy = True
def main():
f1 = torch.randn(1, 64, 64, 64)
block = RepVGGBlock(in_channels=64, out_channels=64)
block.eval()
with torch.no_grad():
output1 = block(f1)
start_time = time.time()
for _ in range(100):
block(f1)
print(f"consume time: {time.time() - start_time}")
# re-parameterization
block.switch_to_deploy()
output2 = block(f1)
start_time = time.time()
for _ in range(100):
block(f1)
print(f"consume time: {time.time() - start_time}")
np.testing.assert_allclose(output1.numpy(), output2.numpy(), rtol=1e-03, atol=1e-05)
print("convert module has been tested, and the result looks good!")
if __name__ == '__main__':
main()