使用python进行贝叶斯统计分析
本文讲解了使用PyMC3进行基本的贝叶斯统计分析过程.
最近我们被客户要求撰写关于贝叶斯统计分析的研究报告,包括一些图形和统计输出。
# 导入
import pymc3 as pm # python的概率编程包
import numpy.random as npr # numpy是用来做科学计算的
import matplotlib.pyplot as plt # matplotlib是用来画图的
import matplotlib as mpl
贝叶斯公式
常见的统计分析问题
- 参数估计: "真实值是否等于X"
- 比较两组实验数据: "实验组是否与对照组不同? "
问题1: 参数估计
"真实值是否等于X?"
或者说
"给定数据,对于感兴趣的参数,可能值的概率分布是多少?"
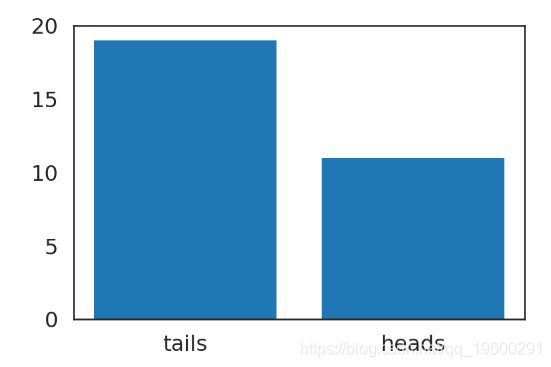
例 1: 抛硬币问题
我把我的硬币抛了 n次,正面是 h次。 这枚硬币是有偏的吗?
参数估计问题parameterized problem
先验假设
- 对参数预先的假设分布: p∼Uniform(0,1)
- likelihood function(似然函数, 翻译这词还不如英文原文呢): data∼Bernoulli(p)
# 产生所需要的数据
from random import shuffle
total = 30
n_heads = 11
n_tails = total - n_heads
tosses = [1] * n_heads + [0] * n_tails
shuffle(tosses)
数据
fig = plot_coins()
plt.show()
MCMC Inference Button (TM)
100%|██████████| 2500/2500 [00:00<00:00, 3382.23it/s]
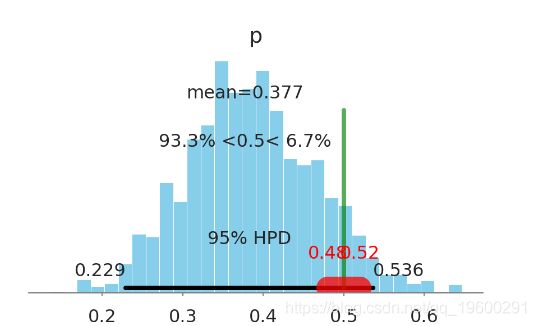
结果
pm.traceplot(coin_trace)
plt.show()
In [10]:
plt.show()
- 95% highest posterior density (HPD, 大概类似于置信区间) 包含了 region of practical equivalence (ROPE, 实际等同区间).
例 2: 药品问题
我有一个新开发的X; X在阻止流感病毒复制方面有多好?
实验
-
测试X的浓度范围, 测量流感活性
-
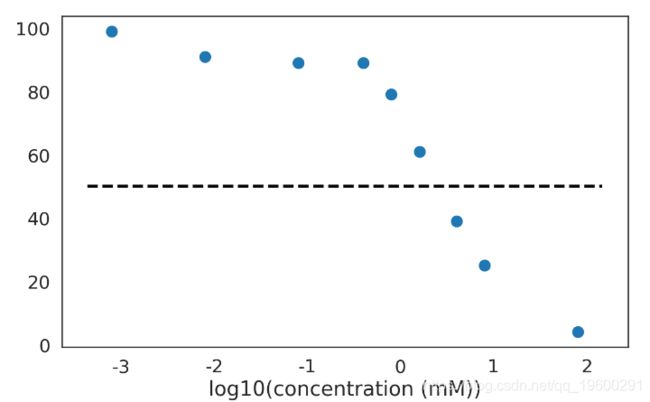
计算 IC50: 能够抑制病毒复制活性50%的X浓度.
data
import pandas as pd
chem_df = pd.DataFrame(chem_data)
chem_df.columns = ['concentration', 'activity']
chem_df['concentration_log'] = chem_df['concentration'].apply(lambda x:np.log10(x))
# df.set_index('concentration', inplace=True)
参数化问题parameterized problem
给定数据, 求出化学物质的IC50值是多少, 并且求出置信区间( 原文中the uncertainty surrounding it, 后面看类似置信区间的含义)?
先验知识
- 由药学知识已知测量函数(measurement function): m=β1+ex−IC50
- 测量函数中的参数估计, 来自先验知识: β∼HalfNormal(1002)
- 关于感兴趣参数的先验知识: log(IC50)∼ImproperFlat
- likelihood function: data∼N(m,1)
数据
In [13]:
fig = plot_chemical_data(log=True)
plt.show()
MCMC Inference Button (TM)
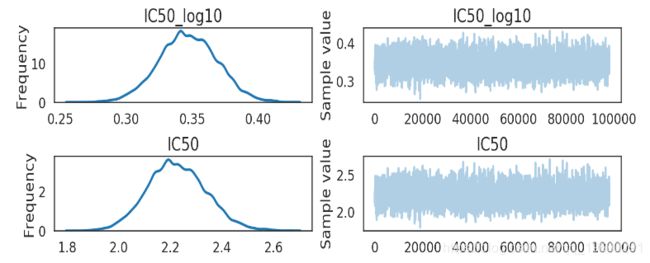
In [16]:
pm.traceplot(ic50_trace[2000:], varnames=['IC50_log10', 'IC50']) # 实时:从步骤2000开始的示例。
plt.show()
结果
In [17]:
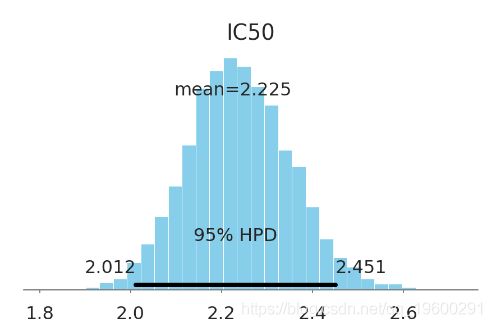
pm.plot_posterior(ic50_trace[4000:], varnames=['IC50'],
color='#87ceeb', point_estimate='mean')
plt.show()
该化学物质的 IC50 大约在[2 mM, 2.4 mM] (95% HPD). 这不是个好的药物候选者.
第二类问题: 实验组之间的比较
"实验组和对照组之间是否有差别? "
例 1: 药品对IQ的影响问题
药品治疗是否影响(提高)IQ分数?
def ECDF(data):
x = np.sort(data)
y = np.cumsum(x) / np.sum(x)
return x, y
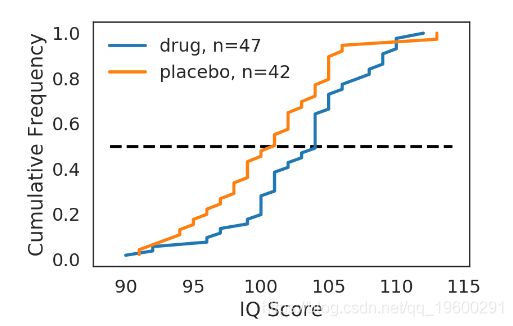
def plot_drug():
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
x_drug, y_drug = ECDF(drug)
ax.plot(x_drug, y_drug, label='drug, n={0}'.format(len(drug)))
x_placebo, y_placebo = ECDF(placebo)
ax.plot(x_placebo, y_placebo, label='placebo, n={0}'.format(len(placebo)))
ax.legend()
ax.set_xlabel('IQ Score')
ax.set_ylabel('Cumulative Frequency')
ax.hlines(0.5, ax.get_xlim()[0], ax.get_xlim()[1], linestyle='--')
return fig
Out[19]:
Ttest_indResult(statistic=2.2806701634329549, pvalue=0.025011500508647616)实验
- 参与者被随机分为两组:
治疗组vs.安慰剂组
- 测量参与者的IQ分数
先验知识
- 被测数据符合t分布: data∼StudentsT(μ,σ,ν)
以下为t分布的几个参数:
- 均值符合正态分布: μ∼N(0,1002)
- 自由度(degrees of freedom)符合指数分布: ν∼Exp(30)
- 方差是positively-distributed: σ∼HalfCauchy(1002)
数据
In [20]:
fig = plot_drug()
plt.show()
代码
In [21]:
y_vals = np.concatenate([drug, placebo])
labels = ['drug'] * len(drug) + ['placebo'] * len(placebo)
data = pd.DataFrame([y_vals, labels]).T
data.columns = ['IQ', 'treatment']
MCMC Inference Button (TM)
结果
In [24]:
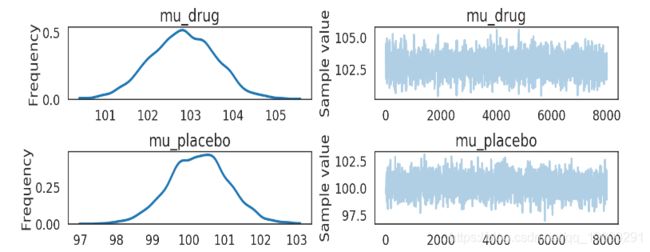
pm.traceplot(kruschke_trace[2000:],
varnames=['mu_drug', 'mu_placebo'])
plt.show()
In [25]:
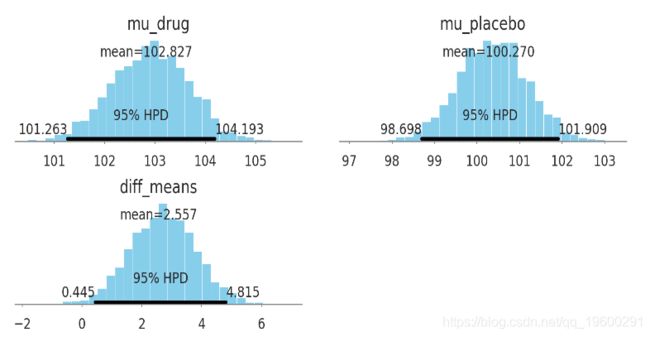
pm.plot_posterior(kruschke_trace[2000:], color='#87ceeb',
varnames=['mu_drug', 'mu_placebo', 'diff_means'])
plt.show()
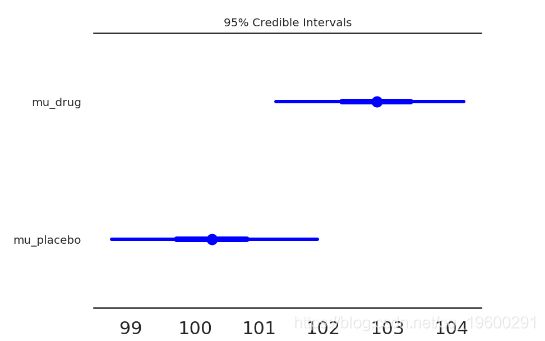
- IQ均值的差距为: [0.5, 4.6]
- 频率主义的 p-value: 0.02 (!!!!!!!!)
注: IQ的差异在10以上才有点意义. p-value=0.02说明组间有差异, 但没说差异有多大..
In [27]:
ax = adjust_forestplot_for_slides(ax)
plt.show()
森林图:在同一轴上的95%HPD(细线),IQR(粗线)和后验分布的中位数(点),使我们能够直接比较治疗组和对照组。
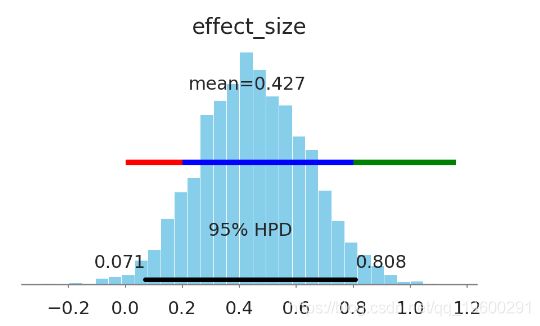
In [29]:
ax = pm.plot_posterior(kruschke_trace[2000:],
varnames=['effect_size'],
color='#87ceeb')
overlay_effect_size(ax)
- 这种药很可能是无关紧要的。
- 没有生物学意义的证据。
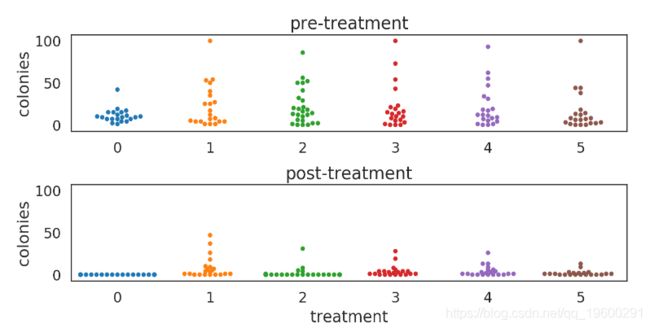
例 2: 手机消毒问题
比较两种常用的消毒方法, 哪种消毒方法更好
实验设计
- 将手机随机分到6组: 4 "fancy" 方法 + 2 "control" 方法.
- 处理前后对手机表面进行拭子菌培养
- count 菌落数量, 比较处理前后的菌落计数
Out[30]:
sample_id int32
treatment int32
colonies_pre int32
colonies_post int32
morphologies_pre int32
morphologies_post int32
year float32
month float32
day float32
perc_reduction morph float32
site int32
phone ID float32
no case float32
frac_change_colonies float64
dtype: object数据
In [32]:
fig = plot_colonies_data()
plt.show()
先验知识
菌落计数符合泊松Poisson分布.
- 菌落计数符合泊松分布: dataij∼Poisson(μij),j∈[pre,post],i∈[1,2,3...]
- 泊松分布的参数是离散均匀分布: μij∼DiscreteUniform(0,104),j∈[pre,post],i∈[1,2,3...]
- 灭菌效力通过百分比变化测量,定义如下: mupre−mupostmupre
MCMC Inference Button (TM)
In [34]:
with poisson_estimation:
poisson_trace = pm.sample(200000)
Assigned Metropolis to pre_mus
Assigned Metropolis to post_mus
100%|██████████| 200500/200500 [01:15<00:00, 2671.98it/s]
In [35]:
pm.traceplot(poisson_trace[50000:], varnames=['pre_mus', 'post_mus'])
plt.show()
结果
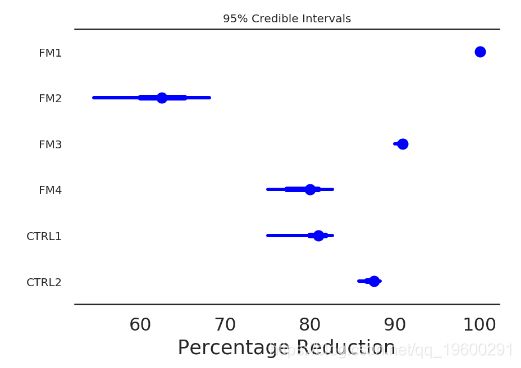
In [39]:
pm.forestplot(poisson_trace[50000:], varnames=['perc_change'],
ylabels=treatment_order) #, xrange=[0, 110])
plt.xlabel('Percentage Reduction')
ax = plt.gca()
ax = adjust_forestplot_for_slides(ax)