如何使用scipy.stats快速对统计量拟合多种统计分布并进行可视化
由于我们在对统计量统计频率后,往往需要对统计量拟合其合适的分布,但scipy.stats提供了超过80种的统计分布,我们不可能手工对其进行尝试,因此以下代码将选择多种常用的统计分布快速对统计量进行拟合,并通过卡方检验返回其最适合的3种分布以及其对应的参数,最后将统计量的分布与其拟合的分布进行可视化。
1. 载入数据,并将数据进行array化
import pandas as pd
import numpy as np
import scipy
from sklearn.preprocessing import StandardScaler
import scipy.stats
import matplotlib.pyplot as plt
%matplotlib inline
# Load data and select first column
from sklearn import datasets
y=np.array(df_test.week)
# Create an index array (x) for data
x = np.arange(len(y))
size = len(y)
2. 进行统计量的分布展示
plt.hist(y)
plt.show()
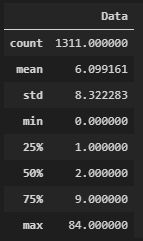
3. 展示统计量的统计概况
y_df = pd.DataFrame(y, columns=['Data'])
y_df.describe()
4. 对数据进行标准化
sc=StandardScaler()
yy = y.reshape (-1,1)
sc.fit(yy)
y_std =sc.transform(yy)
y_std = y_std.flatten()
y_std
del yy
5. 快速拟合10种分布并进行卡方检验,再从高到低返回最合适的分布,卡方值和p值,最后用最大似然函数返回最合适的3种分布所对应的参数。
注意:
由于卡方检验中,在计算期望频率的数组中可能会出现NaN,如果不进行处理,将使得累计的期望频率和(cum_expected_frequency)也计算为NaN,进而导致卡方检验值为空,故在这里需要对NaN进行替换,因此可考虑填0来替换NaN值 (在下列代码中有进行标注)。
# Set list of distributions to test
# See https://docs.scipy.org/doc/scipy/reference/stats.html for more
# Turn off code warnings (this is not recommended for routine use)
import warnings
warnings.filterwarnings("ignore")
# Set up list of candidate distributions to use
# See https://docs.scipy.org/doc/scipy/reference/stats.html for more
dist_names = ['beta',
'expon',
'gamma',
'lognorm',
'norm',
'pearson3',
'triang',
'uniform',
'weibull_min',
'weibull_max']
# Set up empty lists to stroe results
chi_square = []
p_values = []
# Set up 50 bins for chi-square test
# Observed data will be approximately evenly distrubuted aross all bins
percentile_bins = np.linspace(0,100,51)
percentile_cutoffs = np.percentile(y_std, percentile_bins)
observed_frequency, bins = (np.histogram(y_std, bins=percentile_cutoffs))
cum_observed_frequency = np.cumsum(observed_frequency)
# Loop through candidate distributions
for distribution in dist_names:
# Set up distribution and get fitted distribution parameters
dist = getattr(scipy.stats, distribution)
param = dist.fit(y_std)
# Obtain the KS test P statistic, round it to 5 decimal places
p = scipy.stats.kstest(y_std, distribution, args=param)[1]
p = np.around(p, 5)
p_values.append(p)
# Get expected counts in percentile bins
# This is based on a 'cumulative distrubution function' (cdf)
cdf_fitted = dist.cdf(percentile_cutoffs, *param[:-2], loc=param[-2],
scale=param[-1])
expected_frequency = []
for bin in range(len(percentile_bins)-1):
expected_cdf_area = cdf_fitted[bin+1] - cdf_fitted[bin]
expected_frequency.append(expected_cdf_area)
# calculate chi-squared
expected_frequency = np.array(expected_frequency) * size
cum_expected_frequency = np.cumsum(expected_frequency)
# 注意:由于cum_expected_frequency的数组中可能会出现NaN,如果不替换NaN,将无法返回卡方的结果,故在这里需要对NaN进行替换,我这里替换的方式是填0。
l = ((cum_expected_frequency - cum_observed_frequency) ** 2) / cum_observed_frequency
l[np.isnan(l)] = 0
ss = sum (((cum_expected_frequency - cum_observed_frequency) ** 2) / cum_observed_frequency)
chi_square.append(ss)
# Collate results and sort by goodness of fit (best at top)
results = pd.DataFrame()
results['Distribution'] = dist_names
results['chi_square'] = chi_square
results['p_value'] = p_values
results.sort_values(['chi_square'], inplace=True)
# Report results
print ('\nDistributions sorted by goodness of fit:')
print ('----------------------------------------')
print (results)
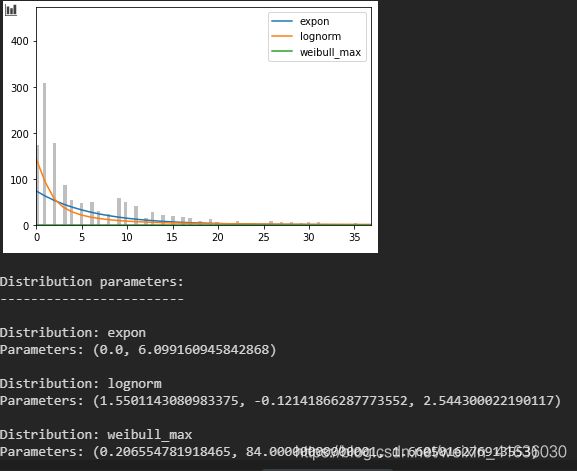
6. 将前3种分布与统计量进行拟合画图,并返回对应的参数值
# Divide the observed data into 100 bins for plotting (this can be changed)
number_of_bins = 100
bin_cutoffs = np.linspace(np.percentile(y,0), np.percentile(y,99),number_of_bins)
# Create the plot
h = plt.hist(y, bins = bin_cutoffs, color='0.75')
# Get the top three distributions from the previous phase
number_distributions_to_plot = 3
dist_names = results['Distribution'].iloc[0:number_distributions_to_plot]
# Create an empty list to stroe fitted distribution parameters
parameters = []
# Loop through the distributions ot get line fit and paraemters
for dist_name in dist_names:
# Set up distribution and store distribution paraemters
dist = getattr(scipy.stats, dist_name)
param = dist.fit(y)
parameters.append(param)
# Get line for each distribution (and scale to match observed data)
pdf_fitted = dist.pdf(x, *param[:-2], loc=param[-2], scale=param[-1])
scale_pdf = np.trapz (h[0], h[1][:-1]) / np.trapz (pdf_fitted, x)
pdf_fitted *= scale_pdf
# Add the line to the plot
plt.plot(pdf_fitted, label=dist_name)
# Set the plot x axis to contain 99% of the data
# This can be removed, but sometimes outlier data makes the plot less clear
plt.xlim(0,np.percentile(y,99))
# Add legend and display plot
plt.legend()
plt.show()
# Store distribution paraemters in a dataframe (this could also be saved)
dist_parameters = pd.DataFrame()
dist_parameters['Distribution'] = (
results['Distribution'].iloc[0:number_distributions_to_plot])
dist_parameters['Distribution parameters'] = parameters
# Print parameter results
print ('\nDistribution parameters:')
print ('------------------------')
for index, row in dist_parameters.iterrows():
print ('\nDistribution:', row[0])
print ('Parameters:', row[1] )
结果展示:
由下图可知,指数分布表现最好,且通过MLE估计出其参数为(0.0, 6.01)。
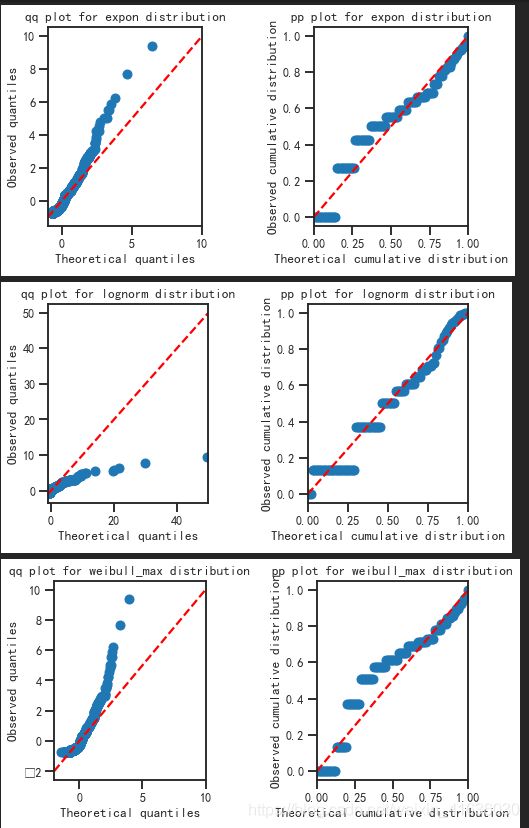
7. 用qq和pp plot去检验该分布有多适合当前的统计量
## qq and pp plots
data = y_std.copy()
data.sort()
# Loop through selected distributions (as previously selected)
for distribution in dist_names:
# Set up distribution
dist = getattr(scipy.stats, distribution)
param = dist.fit(y_std)
# Get random numbers from distribution
norm = dist.rvs(*param[0:-2],loc=param[-2], scale=param[-1],size = size)
norm.sort()
# Create figure
fig = plt.figure(figsize=(8,5))
# qq plot
ax1 = fig.add_subplot(121) # Grid of 2x2, this is suplot 1
ax1.plot(norm,data,"o")
min_value = np.floor(min(min(norm),min(data)))
max_value = np.ceil(max(max(norm),max(data)))
ax1.plot([min_value,max_value],[min_value,max_value],'r--')
ax1.set_xlim(min_value,max_value)
ax1.set_xlabel('Theoretical quantiles')
ax1.set_ylabel('Observed quantiles')
title = 'qq plot for ' + distribution +' distribution'
ax1.set_title(title)
# pp plot
ax2 = fig.add_subplot(122)
# Calculate cumulative distributions
bins = np.percentile(norm,range(0,101))
data_counts, bins = np.histogram(data,bins)
norm_counts, bins = np.histogram(norm,bins)
cum_data = np.cumsum(data_counts)
cum_norm = np.cumsum(norm_counts)
cum_data = cum_data / max(cum_data)
cum_norm = cum_norm / max(cum_norm)
# plot
ax2.plot(cum_norm,cum_data,"o")
min_value = np.floor(min(min(cum_norm),min(cum_data)))
max_value = np.ceil(max(max(cum_norm),max(cum_data)))
ax2.plot([min_value,max_value],[min_value,max_value],'r--')
ax2.set_xlim(min_value,max_value)
ax2.set_xlabel('Theoretical cumulative distribution')
ax2.set_ylabel('Observed cumulative distribution')
title = 'pp plot for ' + distribution +' distribution'
ax2.set_title(title)
# Display plot
plt.tight_layout(pad=4)
plt.show()
结果展示:
从下图的qqplot可以看出,指数分布将更加适合当前的统计量
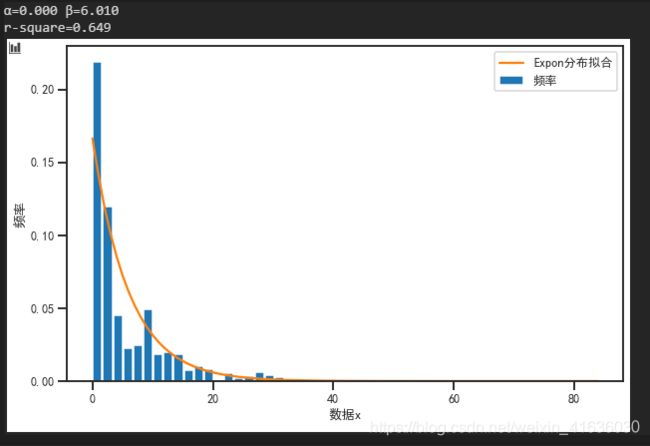
8. 对统计量与拟合的概率分布进行可视化对比
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as st
import numpy as np
class Exponfitplot:
def __init__(self, x, num_bins, legends=None, labels=None, fsize=(10, 6.18), show_info=True):
#:param x: 数据x列表
#:param num_bins: x的分组数
#:param legends: 图例名,默认为 "频率", "Expon分布拟合"
#:param labels:坐标轴标题名,默认为 "数据x", "数据y"
#:param show_info:是否显示拟合结果信息
if legends is None:
legends = ["频率", "Expon分布拟合"]
if labels is None:
labels = ["数据x", "频率"]
self.x = x
self.num_bins = num_bins
self.paras = (0, 6.01)
self.fsize = fsize
self.legends = legends
self.labels = labels
self.show_info = show_info
def change_legend(self, new_legends):
# 更改图例
self.legends = new_legends
def change_label(self, new_labels):
# 更改坐标轴标题
self.labels = new_labels
def change_para(self, new_paras):
# 更改gamma分布的参数
self.paras = new_paras
def draw_plot(self):
fs = self.fsize # 画布大小
# alpha = self.paras[0] # gamma分布中的参数α
# beta = self.paras[1] # gamma分布中的参数β
# 利用seaborn库对字体大小进行统一设置,为fgsize[1]的0.12倍,即画布纵向大小为1000时,font_scale=1.2
sns.set_style("ticks")
sns.set_context("talk", font_scale=fs[1]*0.12)
plt.rcParams['font.sans-serif'] = ['SimHei']
# 设置画布
plt.figure(figsize=fs)
# ax:绘制频率直方图
plt.hist(self.x, self.num_bins, density=1, rwidth=0.9, label=self.legends[0])
plt.xlabel(self.labels[0])
plt.ylabel(self.labels[1])
# ax2:绘制gamma拟合曲线
self.x.sort()
y = st.expon.pdf(self.x, 0, scale=6.01)
plt.plot(self.x, y, '-', label=self.legends[1])
if self.show_info:
info = "α={:.3f} β={:.3f}\nr-square={:.3f}".format(self.paras[0], self.paras[1], st.linregress(self.x, y)[2]**2)
print(info)
# 显示多图例
plt.legend(loc=1, bbox_to_anchor=(1, 1))
if __name__ == "__main__":
np.random.seed(19680801)
iqs = np.array(df_test.week) # 待绘制直方图的数据
bins = 50 # 数据分组数
gammaplot = Exponfitplot(iqs, bins)
gammaplot.draw_plot()
plt.show()
结果展示:
从下图中可以看出,expon(x, loc= 0, scale = 6.01) 的指数分布对于该统计量的拟合效果比较好
9. 校验:定义该指数函数,并根据统计量的值返回对应的指数分布的概率
f_0 = scipy.stats.expon.pdf(0, loc = 0, scale = 6.01)
print('the probablity of exponential distribution is {0} when x = 0'.format(f_0))
结果展示:
![]()