三、生成heatmap(一)裁背景和组织patch
提出来的新思路:
将按顺序裁剪的patch喂入已训练好的网络,所生成的概率重新赋给数组中的对应区域,生成热力图。
某张tif及所画病灶
if __name__ == '__main__':
wsi = WSI() # 创建一个wsi对象
# 含read_wsi_tumor()、find_roi_n_extract_patches_tumor()、extract_patches_tumor()
run_on_tumor_data() #见↓↓↓↓↓↓def run_on_tumor_data():
wsi.wsi_paths = glob.glob(os.path.join(utils.TEST_TUMOR_WSI_PATH, '*.tif'))
wsi.wsi_paths.sort() # 读原图.tif
wsi.mask_paths = glob.glob(os.path.join(utils.TEST_TUMOR_MASK_PATH, '*.tif'))
wsi.mask_paths.sort() # 读mask.tif
wsi.index = 0
for wsi_path, mask_path in zip(wsi.wsi_paths, wsi.mask_paths):
if wsi.read_wsi_tumor(wsi_path, mask_path): #见下{1}
wsi.find_roi_n_extract_patches_tumor() #见下{2}{1} 函数 read_wsi_tumor()
def read_wsi_tumor(self, wsi_path, mask_path):
try:
self.cur_wsi_path = wsi_path
self.wsi_image = OpenSlide(wsi_path) # 读入原图wsi_image
self.mask_image = OpenSlide(mask_path) # 读入掩膜mask_image
# 所用的level
self.level_used = min(self.def_level, self.wsi_image.level_count - 1, self.mask_image.level_count - 1)
# 读所用的level的切片图
self.rgb_image_pil = self.wsi_image.read_region((0, 0), self.level_used,
self.wsi_image.level_dimensions[self.level_used])
self.rgb_image = np.array(self.rgb_image_pil)
except OpenSlideUnsupportedFormatError:
print('Exception: OpenSlideUnsupportedFormatError')
return False
return True{2} 函数 find_roi_n_extract_patches_tumor()
def find_roi_n_extract_patches_tumor(self):
[1] hsv = cv2.cvtColor(self.rgb_image, cv2.COLOR_BGR2HSV)
lower_red = np.array([20, 20, 20])
upper_red = np.array([255, 255, 255])
mask = cv2.inRange(hsv, lower_red, upper_red)
# (50, 50)
[2] close_kernel = np.ones((50, 50), dtype=np.uint8)
# 形态学滤波,具体见下
image_close = Image.fromarray(cv2.morphologyEx(np.array(mask), cv2.MORPH_CLOSE, close_kernel))
# (30, 30)
open_kernel = np.ones((30, 30), dtype=np.uint8)
# 形态学滤波,具体见下
image_open = Image.fromarray(cv2.morphologyEx(np.array(image_close), cv2.MORPH_OPEN, open_kernel))
[3] contour_rgb, bounding_boxes = self.get_image_contours_tumor(np.array(image_open), self.rgb_image)
#bounding_boxes[0]=x_start [1]=y_start [3]=x_length [4]=y_length
#contour_rgb = cv2.resize(contour_rgb, (0, 0), fx=0.40, fy=0.40)
#cv2.imshow('contour_rgb', np.array(contour_rgb))
self.rgb_image_pil.close()
[4] self.extract_patches_tumor(bounding_boxes)
self.wsi_image.close()
self.mask_image.close()
/// [1] 基于HSV的阈值分割
HSV的阈值如何得出:通过debug→hsv = cv2.cvtColor(self.rgb_image, cv2.COLOR_BGR2HSV)
选了三种背景看HSV,发现S值很小(<20),通过公式计算也可以得出范围
/// [2] 开、闭操作
属于针对二值图像的形态学滤波
- 腐蚀操作:(目标变小)
将图像外围的突出点加以腐蚀,核与其覆盖的图像部分做“与”操作,如果全为1,则该像素点为1,否则为0;也就是0容易得到,图像更多的地方变黑了,白色部分被腐蚀了。
- 膨胀操作:(目标变大)
与腐蚀相反,核与其覆盖的图像部分做“或”操作,如果全为0,则该像素点为0,否则为1;也就是1容易得到,图像更多的地方变白了,白色部分膨胀了。
- 闭操作(先膨胀,再腐蚀)
排除小型背景黑洞,可以将许多靠近的图块相连称为一个无突起的连通域
- 开操作(先腐蚀,再膨胀)
作用:放大裂缝和低密度区域,消除小目标物体,在平滑较大物体的边界时,不改变其面积。消除物体表面的突起。
为什么是先闭后开:可以试试先开后闭的效果
/// [3] get_image_contours_tumor()
def get_image_contours_tumor(cont_img, rgb_image):
contours, _ = cv2.findContours(cont_img, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
# 对cont_img只检测外轮廓,矩形轮廓只需4个点来保存。返回轮廓本身
bounding_boxes = [cv2.boundingRect(c) for c in contours]
contours_rgb_image_array = np.array(rgb_image)
line_color = (255, 0, 0) # blue color code
cv2.drawContours(contours_rgb_image_array, contours, -1, line_color, 3)
# 所画图片,cv2.findContours()找出来的轮廓点集,全部绘制可设为-1,轮廓的颜色和厚度
return contours_rgb_image_array, bounding_boxes/// [4] extract_patches_tumor()
def extract_patches_tumor(self, bounding_boxes):
mag_factor = pow(2, self.level_used) # 对应放大倍数
print('No. of ROIs to extract patches from: %d' % len(bounding_boxes))
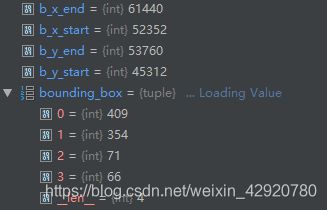
#bounding_boxes[0]=x_start [1]=y_start [3]=x_length [4]=y_length
for i, bounding_box in enumerate(bounding_boxes):
b_x_start = int(bounding_box[0]) * mag_factor
b_y_start = int(bounding_box[1]) * mag_factor
b_x_end = (int(bounding_box[0]) + int(bounding_box[2])) * mag_factor
b_y_end = (int(bounding_box[1]) + int(bounding_box[3])) * mag_factor
X = np.arange(b_x_start, b_x_end, 256) # X隔256取一个数
Y = np.arange(b_y_start, b_y_end, 256) # Y隔256取一个数
for x in X :
for y in Y :
patch = self.wsi_image.read_region((x, y), 0, (utils.PATCH_SIZE, utils.PATCH_SIZE))
mask = self.mask_image.read_region((x, y), 0, (utils.PATCH_SIZE, utils.PATCH_SIZE))
mask_gt = np.array(mask)
mask_gt = cv2.cvtColor(mask_gt, cv2.COLOR_BGR2GRAY) # 二值化
patch_array = np.array(patch)
white_pixel_cnt_gt = cv2.countNonZero(mask_gt)
# 若有病区域面积>0.85,则positive
if white_pixel_cnt_gt >= ((utils.PATCH_SIZE * utils.PATCH_SIZE) * 0.85):
patch.save(utils.PATCHES_TEST_POSITIVE_PATH + str(self.patch_index) + '_y.PNG')
else:
patch_hsv = cv2.cvtColor(patch_array, cv2.COLOR_BGR2HSV)
lower_red = np.array([20, 20, 20])
upper_red = np.array([200, 200, 200])
mask_patch = cv2.inRange(patch_hsv, lower_red, upper_red)
white_pixel_cnt = cv2.countNonZero(mask_patch) # 组织
# 若ROI面积>0.5,则negative,否则为背景
if white_pixel_cnt > ((utils.PATCH_SIZE * utils.PATCH_SIZE) * 0.5):
patch.save(utils.PATCHES_TEST_NEGATIVE_PATH + str(self.patch_index) + '_y.PNG')
else:
patch.save(utils.PATCHES_TEST_BACKGROUND_PATH + str(self.patch_index) + '_n.PNG')
self.patch_index += 1
print(x, y)
patch.close()
mask.close()分割结果: