SparkML之分类(一)贝叶斯分类
1.1、贝叶斯定理
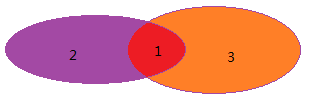
贝叶斯定理:用来描述两个条件概率之间的关系。比如P(A/B)和P(B/A),那么可以推导:![]() ,我们下图进行进行说明:
,我们下图进行进行说明:
那么有贝叶斯定理公式:
1.2、朴素贝叶斯分类器( Naive Bayes Classifiers)
大家知道最为广泛的两个分类模型就是决策树模型和朴素贝叶斯分类模型,前者是对象属性与对象值之间的一种映射关系,后者则是用那个概率最大,那么待分类项就属于哪个类别(对象属性和概率值之间的一种映射)。
源码的理论链接为:
/**
* Trains a Naive Bayes model given an RDD of `(label, features)` pairs.
*
* This is the Multinomial NB ([[http://tinyurl.com/lsdw6p]]) which can handle all kinds of
* discrete data. For example, by converting documents into TF-IDF vectors, it can be used for
* document classification. By making every vector a 0-1 vector, it can also be used as
* Bernoulli NB ([[http://tinyurl.com/p7c96j6]]). The input feature values must be nonnegative.
*/下面简单陈述一下它的思想
這个是最基本的贝叶斯分类的思想,在结合其他领域情况下,在计算概率P這快有所不同,在spark源码中的计算 概率P是在NaiveBayes這个类下面的run方法下,源码如下:
def run(data: RDD[LabeledPoint]): NaiveBayesModel = {
val requireNonnegativeValues: Vector => Unit = (v: Vector) => {
val values = v match {
case sv: SparseVector => sv.values
case dv: DenseVector => dv.values
}
if (!values.forall(_ >= 0.0)) {
throw new SparkException(s"Naive Bayes requires nonnegative feature values but found $v.")
}
}
val requireZeroOneBernoulliValues: Vector => Unit = (v: Vector) => {
val values = v match {
case sv: SparseVector => sv.values
case dv: DenseVector => dv.values
}
if (!values.forall(v => v == 0.0 || v == 1.0)) {
throw new SparkException(
s"Bernoulli naive Bayes requires 0 or 1 feature values but found $v.")
}
}
// 对每个标签进行聚合操作(Aggregates term frequencies per label).
// TODO: Calling combineByKey and collect creates two stages, we can implement something
// TODO: similar to reduceByKeyLocally to save one stage.
val aggregated = data.map(p => (p.label, p.features)).combineByKey[(Long, DenseVector)](
//检测方法
createCombiner = (v: Vector) => {
if (modelType == Bernoulli) {
requireZeroOneBernoulliValues(v)
} else {
requireNonnegativeValues(v)
}
(1L, v.copy.toDense)
},
mergeValue = (c: (Long, DenseVector), v: Vector) => {
requireNonnegativeValues(v)
BLAS.axpy(1.0, v, c._2) //c.2 += 1.0*v
(c._1 + 1L, c._2)
},
mergeCombiners = (c1: (Long, DenseVector), c2: (Long, DenseVector)) => {
BLAS.axpy(1.0, c2._2, c1._2) c.2 += 1.0*c2._2
(c1._1 + c2._1, c1._2)
}
).collect().sortBy(_._1)
//标签数量
val numLabels = aggregated.length
//聚合文档数量
var numDocuments = 0L
aggregated.foreach { case (_, (n, _)) =>
numDocuments += n
}
//特征数量
val numFeatures = aggregated.head match { case (_, (_, v)) => v.size }
val labels = new Array[Double](numLabels)
//建立一个Array来存储pi类别的先验概率
val pi = new Array[Double](numLabels)
//特征下的条件概率
val theta = Array.fill(numLabels)(new Array[Double](numFeatures))
val piLogDenom = math.log(numDocuments + numLabels * lambda)
var i = 0
aggregated.foreach { case (label, (n, sumTermFreqs)) =>
labels(i) = label
pi(i) = math.log(n + lambda) - piLogDenom
val thetaLogDenom = modelType match {
case Multinomial => math.log(sumTermFreqs.values.sum + numFeatures * lambda)
case Bernoulli => math.log(n + 2.0 * lambda)
case _ =>
// This should never happen.
throw new UnknownError(s"Invalid modelType: $modelType.")
}
var j = 0
while (j < numFeatures) {
theta(i)(j) = math.log(sumTermFreqs(j) + lambda) - thetaLogDenom
j += 1
}
i += 1
}
//返回一个NaiveBayesModel,這个模型输入参数包含 标签labels,先验概率pi,条件概率theta ,模型方法 modelType(仅支持Multinomial、Bernoulli )
new NaiveBayesModel(labels, pi, theta, modelType)
}
} val piLogDenom = math.log(numDocuments + numLabels * lambda)
pi(i) = math.log(n + lambda) - piLogDenom* @param pi log of class priors, whose dimension is C, number of labels那么对于类别集合C = {y1,y2,...,yo}它的先验概率是:
lamda是平滑因子
numDoucuments:总的次数,犹如上面历史数据下男和女的总人数
numlaebls:类别数,犹如上面就之后男女两个类别
再看
theta(i)(j) = math.log(sumTermFreqs(j) + lambda) - thetaLogDenom其中 thetaLogDenom有两种模式
(1)多项式模式:
case Multinomial => math.log(sumTermFreqs.values.sum + numFeatures * lambda)sumTermFreqs.values.sum为解释类别 yi 的总数
(2)伯努利模式
case Bernoulli => math.log(n + 2.0 * lambda)
那么theta(i)(j):
发现当 C的类别数量为 2时,两个公式是相等的
Spark源码
/**
* 朴素贝叶斯分类器模型
*
* @param labels 标签
* @param pi 对先验概率取log之后的值,维度是类别集合C的维度一样
* @param theta 条件概率取log,它的维度是C-by-D D是特征数
* @param modelType 模型的模式类型,选择可以是 "multinomial" or "bernoulli"
*/
@Since("0.9.0")
class NaiveBayesModel private[spark] (
@Since("1.0.0") val labels: Array[Double],
@Since("0.9.0") val pi: Array[Double],
@Since("0.9.0") val theta: Array[Array[Double]],
@Since("1.4.0") val modelType: String)
extends ClassificationModel with Serializable with Saveable {
import NaiveBayes.{Bernoulli, Multinomial, supportedModelTypes}
private val piVector = new DenseVector(pi)
private val thetaMatrix = new DenseMatrix(labels.length, theta(0).length, theta.flatten, true)
private[mllib] def this(labels: Array[Double], pi: Array[Double], theta: Array[Array[Double]]) =
this(labels, pi, theta, NaiveBayes.Multinomial)
/** A Java-friendly constructor that takes three Iterable parameters. */
private[mllib] def this(
labels: JIterable[Double],
pi: JIterable[Double],
theta: JIterable[JIterable[Double]]) =
this(labels.asScala.toArray, pi.asScala.toArray, theta.asScala.toArray.map(_.asScala.toArray))
require(supportedModelTypes.contains(modelType),
s"Invalid modelType $modelType. Supported modelTypes are $supportedModelTypes.")
// Bernoulli 概率得分(大小)要求,当为1时,那么为log(condprob),当为0时,那么为log(1-condprob)
// 這个预计算的log(1.0 - exp(theta))和它们的和为的是应用于這种情况下的预测
//This precomputes log(1.0 - exp(theta)) and its sum which are used for the linear algebra
// application of this condition (in predict function).
private val (thetaMinusNegTheta, negThetaSum) = modelType match {
case Multinomial => (None, None)
case Bernoulli =>
val negTheta = thetaMatrix.map(value => math.log(1.0 - math.exp(value)))
val ones = new DenseVector(Array.fill(thetaMatrix.numCols) {1.0})
val thetaMinusNegTheta = thetaMatrix.map { value =>
value - math.log(1.0 - math.exp(value))
}
(Option(thetaMinusNegTheta), Option(negTheta.multiply(ones)))
case _ =>
// This should never happen.
throw new UnknownError(s"Invalid modelType: $modelType.")
}
@Since("1.0.0")
override def predict(testData: RDD[Vector]): RDD[Double] = {
val bcModel = testData.context.broadcast(this)
testData.mapPartitions { iter =>
val model = bcModel.value
iter.map(model.predict)
}
}
@Since("1.0.0")
override def predict(testData: Vector): Double = {
modelType match {
case Multinomial =>
labels(multinomialCalculation(testData).argmax)
case Bernoulli =>
labels(bernoulliCalculation(testData).argmax)
}
}
/**
* 输入数据,根据模型,进行预测
*
* @param testData 用RDD表示的数据,用于预测
* @return an RDD[Vector] 预测返回值
*/
@Since("1.5.0")
def predictProbabilities(testData: RDD[Vector]): RDD[Vector] = {
val bcModel = testData.context.broadcast(this)
testData.mapPartitions { iter =>
val model = bcModel.value
iter.map(model.predictProbabilities)
}
}
/**
* 使用该模型训练的一个单一数据点的后验概率。
* Predict posterior class probabilities for a single data point using the model trained.
*
* @param testData 用RDD表示的数据,用于预测
* @return an RDD[Vector] 预测返回值
*/
@Since("1.5.0")
def predictProbabilities(testData: Vector): Vector = {
modelType match {
case Multinomial =>
posteriorProbabilities(multinomialCalculation(testData))
case Bernoulli =>
posteriorProbabilities(bernoulliCalculation(testData))
}
}
private def multinomialCalculation(testData: Vector) = {
val prob = thetaMatrix.multiply(testData)
BLAS.axpy(1.0, piVector, prob)
prob
}
private def bernoulliCalculation(testData: Vector) = {
testData.foreachActive((_, value) =>
if (value != 0.0 && value != 1.0) {
throw new SparkException(
s"Bernoulli naive Bayes requires 0 or 1 feature values but found $testData.")
}
)
val prob = thetaMinusNegTheta.get.multiply(testData)
BLAS.axpy(1.0, piVector, prob)
BLAS.axpy(1.0, negThetaSum.get, prob)
prob
}
private def posteriorProbabilities(logProb: DenseVector) = {
val logProbArray = logProb.toArray
val maxLog = logProbArray.max
val scaledProbs = logProbArray.map(lp => math.exp(lp - maxLog))
val probSum = scaledProbs.sum
new DenseVector(scaledProbs.map(_ / probSum))
}
@Since("1.3.0")
override def save(sc: SparkContext, path: String): Unit = {
val data = NaiveBayesModel.SaveLoadV2_0.Data(labels, pi, theta, modelType)
NaiveBayesModel.SaveLoadV2_0.save(sc, path, data)
}
override protected def formatVersion: String = "2.0"
}
@Since("1.3.0")
object NaiveBayesModel extends Loader[NaiveBayesModel] {
import org.apache.spark.mllib.util.Loader._
private[mllib] object SaveLoadV2_0 {
def thisFormatVersion: String = "2.0"
/** Hard-code class name string in case it changes in the future */
def thisClassName: String = "org.apache.spark.mllib.classification.NaiveBayesModel"
/** Model data for model import/export */
case class Data(
labels: Array[Double],
pi: Array[Double],
theta: Array[Array[Double]],
modelType: String)
def save(sc: SparkContext, path: String, data: Data): Unit = {
val sqlContext = SQLContext.getOrCreate(sc)
import sqlContext.implicits._
// Create JSON metadata.
val metadata = compact(render(
("class" -> thisClassName) ~ ("version" -> thisFormatVersion) ~
("numFeatures" -> data.theta(0).length) ~ ("numClasses" -> data.pi.length)))
sc.parallelize(Seq(metadata), 1).saveAsTextFile(metadataPath(path))
// Create Parquet data.
val dataRDD: DataFrame = sc.parallelize(Seq(data), 1).toDF()
dataRDD.write.parquet(dataPath(path))
}
@Since("1.3.0")
def load(sc: SparkContext, path: String): NaiveBayesModel = {
val sqlContext = SQLContext.getOrCreate(sc)
// Load Parquet data.
val dataRDD = sqlContext.read.parquet(dataPath(path))
// Check schema explicitly since erasure makes it hard to use match-case for checking.
checkSchema[Data](dataRDD.schema)
val dataArray = dataRDD.select("labels", "pi", "theta", "modelType").take(1)
assert(dataArray.length == 1, s"Unable to load NaiveBayesModel data from: ${dataPath(path)}")
val data = dataArray(0)
val labels = data.getAs[Seq[Double]](0).toArray
val pi = data.getAs[Seq[Double]](1).toArray
val theta = data.getAs[Seq[Seq[Double]]](2).map(_.toArray).toArray
val modelType = data.getString(3)
new NaiveBayesModel(labels, pi, theta, modelType)
}
}
private[mllib] object SaveLoadV1_0 {
def thisFormatVersion: String = "1.0"
/** Hard-code class name string in case it changes in the future */
def thisClassName: String = "org.apache.spark.mllib.classification.NaiveBayesModel"
/** 模型数据的导入导出 Model data for model import/export */
case class Data(
labels: Array[Double],
pi: Array[Double],
theta: Array[Array[Double]])
def save(sc: SparkContext, path: String, data: Data): Unit = {
val sqlContext = SQLContext.getOrCreate(sc)
import sqlContext.implicits._
//建立一个 JSON文件数据 Create JSON metadata.
val metadata = compact(render(
("class" -> thisClassName) ~ ("version" -> thisFormatVersion) ~
("numFeatures" -> data.theta(0).length) ~ ("numClasses" -> data.pi.length)))
sc.parallelize(Seq(metadata), 1).saveAsTextFile(metadataPath(path))
// 建立一个 Parquet文件数据 Create Parquet data.
val dataRDD: DataFrame = sc.parallelize(Seq(data), 1).toDF()
dataRDD.write.parquet(dataPath(path))
}
def load(sc: SparkContext, path: String): NaiveBayesModel = {
val sqlContext = SQLContext.getOrCreate(sc)
// Load Parquet data.
val dataRDD = sqlContext.read.parquet(dataPath(path))
// Check schema explicitly since erasure makes it hard to use match-case for checking.
checkSchema[Data](dataRDD.schema)
val dataArray = dataRDD.select("labels", "pi", "theta").take(1)
assert(dataArray.length == 1, s"Unable to load NaiveBayesModel data from: ${dataPath(path)}")
val data = dataArray(0)
val labels = data.getAs[Seq[Double]](0).toArray
val pi = data.getAs[Seq[Double]](1).toArray
val theta = data.getAs[Seq[Seq[Double]]](2).map(_.toArray).toArray
new NaiveBayesModel(labels, pi, theta)
}
}
override def load(sc: SparkContext, path: String): NaiveBayesModel = {
val (loadedClassName, version, metadata) = loadMetadata(sc, path)
val classNameV1_0 = SaveLoadV1_0.thisClassName
val classNameV2_0 = SaveLoadV2_0.thisClassName
val (model, numFeatures, numClasses) = (loadedClassName, version) match {
case (className, "1.0") if className == classNameV1_0 =>
val (numFeatures, numClasses) = ClassificationModel.getNumFeaturesClasses(metadata)
val model = SaveLoadV1_0.load(sc, path)
(model, numFeatures, numClasses)
case (className, "2.0") if className == classNameV2_0 =>
val (numFeatures, numClasses) = ClassificationModel.getNumFeaturesClasses(metadata)
val model = SaveLoadV2_0.load(sc, path)
(model, numFeatures, numClasses)
case _ => throw new Exception(
s"NaiveBayesModel.load did not recognize model with (className, format version):" +
s"($loadedClassName, $version). Supported:\n" +
s" ($classNameV1_0, 1.0)")
}
assert(model.pi.length == numClasses,
s"NaiveBayesModel.load expected $numClasses classes," +
s" but class priors vector pi had ${model.pi.length} elements")
assert(model.theta.length == numClasses,
s"NaiveBayesModel.load expected $numClasses classes," +
s" but class conditionals array theta had ${model.theta.length} elements")
assert(model.theta.forall(_.length == numFeatures),
s"NaiveBayesModel.load expected $numFeatures features," +
s" but class conditionals array theta had elements of size:" +
s" ${model.theta.map(_.length).mkString(",")}")
model
}
}
/**
* 输入一个RDD ((label, features)` pairs)来训练朴素贝叶斯模型
*
* This is the Multinomial NB ([[http://tinyurl.com/lsdw6p]]) which can handle all kinds of
* discrete data. For example, by converting documents into TF-IDF vectors, it can be used for
* document classification. By making every vector a 0-1 vector, it can also be used as
* Bernoulli NB ([[http://tinyurl.com/p7c96j6]]). The input feature values must be nonnegative.
*/
@Since("0.9.0")
class NaiveBayes private (
private var lambda: Double,
private var modelType: String) extends Serializable with Logging {
import NaiveBayes.{Bernoulli, Multinomial}
@Since("1.4.0")
def this(lambda: Double) = this(lambda, NaiveBayes.Multinomial)
@Since("0.9.0")
def this() = this(1.0, NaiveBayes.Multinomial)
/** 设置平滑参数(因子)Set the smoothing parameter. Default: 1.0. */
@Since("0.9.0")
def setLambda(lambda: Double): NaiveBayes = {
require(lambda >= 0,
s"Smoothing parameter must be nonnegative but got ${lambda}")
this.lambda = lambda
this
}
/** 得到平滑参数(因子)Get the smoothing parameter. */
@Since("1.4.0")
def getLambda: Double = lambda
/**
* 设置模型的模式类型 ,"multinomial" (default) and "bernoulli".
* Set the model type using a string (case-sensitive).
* Supported options: "multinomial" (default) and "bernoulli".
*/
@Since("1.4.0")
def setModelType(modelType: String): NaiveBayes = {
require(NaiveBayes.supportedModelTypes.contains(modelType),
s"NaiveBayes was created with an unknown modelType: $modelType.")
this.modelType = modelType
this
}
/** 得到模式类型 Get the model type. */
@Since("1.4.0")
def getModelType: String = this.modelType
/**
* Run the algorithm with the configured parameters on an input RDD of LabeledPoint entries.
*
* @param data RDD of [[org.apache.spark.mllib.regression.LabeledPoint]].
*/
def run(data: RDD[LabeledPoint]): NaiveBayesModel = {
val requireNonnegativeValues: Vector => Unit = (v: Vector) => {
val values = v match {
case sv: SparseVector => sv.values
case dv: DenseVector => dv.values
}
if (!values.forall(_ >= 0.0)) {
throw new SparkException(s"Naive Bayes requires nonnegative feature values but found $v.")
}
}
val requireZeroOneBernoulliValues: Vector => Unit = (v: Vector) => {
val values = v match {
case sv: SparseVector => sv.values
case dv: DenseVector => dv.values
}
if (!values.forall(v => v == 0.0 || v == 1.0)) {
throw new SparkException(
s"Bernoulli naive Bayes requires 0 or 1 feature values but found $v.")
}
}
// 对每个标签进行聚合操作(Aggregates term frequencies per label).
// TODO: Calling combineByKey and collect creates two stages, we can implement something
// TODO: similar to reduceByKeyLocally to save one stage.
val aggregated = data.map(p => (p.label, p.features)).combineByKey[(Long, DenseVector)](
//检测方法
createCombiner = (v: Vector) => {
if (modelType == Bernoulli) {
requireZeroOneBernoulliValues(v)
} else {
requireNonnegativeValues(v)
}
(1L, v.copy.toDense)
},
mergeValue = (c: (Long, DenseVector), v: Vector) => {
requireNonnegativeValues(v)
BLAS.axpy(1.0, v, c._2) //c.2 += 1.0*v
(c._1 + 1L, c._2)
},
mergeCombiners = (c1: (Long, DenseVector), c2: (Long, DenseVector)) => {
BLAS.axpy(1.0, c2._2, c1._2) c.2 += 1.0*c2._2
(c1._1 + c2._1, c1._2)
}
).collect().sortBy(_._1)
//标签数量
val numLabels = aggregated.length
//聚合文档数量
var numDocuments = 0L
aggregated.foreach { case (_, (n, _)) =>
numDocuments += n
}
//特征数量
val numFeatures = aggregated.head match { case (_, (_, v)) => v.size }
val labels = new Array[Double](numLabels)
//建立一个Array来存储pi类别的先验概率
val pi = new Array[Double](numLabels)
//特征下的条件概率
val theta = Array.fill(numLabels)(new Array[Double](numFeatures))
val piLogDenom = math.log(numDocuments + numLabels * lambda)
var i = 0
aggregated.foreach { case (label, (n, sumTermFreqs)) =>
labels(i) = label
pi(i) = math.log(n + lambda) - piLogDenom
val thetaLogDenom = modelType match {
case Multinomial => math.log(sumTermFreqs.values.sum + numFeatures * lambda)
case Bernoulli => math.log(n + 2.0 * lambda)
case _ =>
// This should never happen.
throw new UnknownError(s"Invalid modelType: $modelType.")
}
var j = 0
while (j < numFeatures) {
theta(i)(j) = math.log(sumTermFreqs(j) + lambda) - thetaLogDenom
j += 1
}
i += 1
}
//返回一个NaiveBayesModel,這个模型包含 标签labels,先验概率pi,条件概率theta ,模型方法 modelType(仅支持Multinomial、Bernoulli )
new NaiveBayesModel(labels, pi, theta, modelType)
}
}
/**
* Top-level methods for calling naive Bayes.
*/
@Since("0.9.0")
object NaiveBayes {
/** String name for multinomial model type. */
private[spark] val Multinomial: String = "multinomial"
/** String name for Bernoulli model type. */
private[spark] val Bernoulli: String = "bernoulli"
/* Set of modelTypes that NaiveBayes supports */
private[spark] val supportedModelTypes = Set(Multinomial, Bernoulli)
/**
* Trains a Naive Bayes model given an RDD of `(label, features)` pairs.
*
* This is the default Multinomial NB ([[http://tinyurl.com/lsdw6p]]) which can handle all
* kinds of discrete data. For example, by converting documents into TF-IDF vectors, it
* can be used for document classification.
*
* This version of the method uses a default smoothing parameter of 1.0.
*
* @param input RDD of `(label, array of features)` pairs. Every vector should be a frequency
* vector or a count vector.
*/
@Since("0.9.0")
def train(input: RDD[LabeledPoint]): NaiveBayesModel = {
new NaiveBayes().run(input)
}
/**
* Trains a Naive Bayes model given an RDD of `(label, features)` pairs.
*
* This is the default Multinomial NB ([[http://tinyurl.com/lsdw6p]]) which can handle all
* kinds of discrete data. For example, by converting documents into TF-IDF vectors, it
* can be used for document classification.
*
* @param input RDD of `(label, array of features)` pairs. Every vector should be a frequency
* vector or a count vector.
* @param lambda The smoothing parameter
*/
@Since("0.9.0")
def train(input: RDD[LabeledPoint], lambda: Double): NaiveBayesModel = {
new NaiveBayes(lambda, Multinomial).run(input)
}
/**
* Trains a Naive Bayes model given an RDD of `(label, features)` pairs.
*
* The model type can be set to either Multinomial NB ([[http://tinyurl.com/lsdw6p]])
* or Bernoulli NB ([[http://tinyurl.com/p7c96j6]]). The Multinomial NB can handle
* discrete count data and can be called by setting the model type to "multinomial".
* For example, it can be used with word counts or TF_IDF vectors of documents.
* The Bernoulli model fits presence or absence (0-1) counts. By making every vector a
* 0-1 vector and setting the model type to "bernoulli", the fits and predicts as
* Bernoulli NB.
*
* @param input RDD of `(label, array of features)` pairs. Every vector should be a frequency
* vector or a count vector.
* @param lambda The smoothing parameter
*
* @param modelType The type of NB model to fit from the enumeration NaiveBayesModels, can be
* multinomial or bernoulli
*/
@Since("1.4.0")
def train(input: RDD[LabeledPoint], lambda: Double, modelType: String): NaiveBayesModel = {
require(supportedModelTypes.contains(modelType),
s"NaiveBayes was created with an unknown modelType: $modelType.")
new NaiveBayes(lambda, modelType).run(input)
}
}
Spark实验
import org.apache.spark.mllib.classification.{NaiveBayes, NaiveBayesModel} import org.apache.spark.mllib.linalg.Vectors import org.apache.spark.mllib.regression.LabeledPoint import org.apache.spark.{SparkConf, SparkContext} object naiveBayes { def main(args: Array[String]) { val conf = new SparkConf().setAppName("naive Bayes example").setMaster("local") val sc = new SparkContext(conf) val data = sc.textFile("C:\\Users\\alienware\\IdeaProjects\\sparkCore\\data\\mllib\\sample_naive_bayes_data.txt") val parseData = data.map{ line => val parts = line.split(",") LabeledPoint(parts(0).toDouble,Vectors.dense(parts(1).split(" ").map( _.toDouble))) } // Split the data into training and test sets (50% held out for testing) val splitData = parseData.randomSplit(Array(0.5,0.5),seed = 1L) val trainData = splitData(0) val testData = splitData(1) // Train naiveBayesModel val model = NaiveBayes.train(trainData,lambda = 1.0,modelType = "multinomial") val labelsAndPredictions = testData.map(p => (model.predict(p.features),p.label)) labelsAndPredictions.foreach(println) /** (0.0,0.0) (1.0,1.0) (1.0,1.0) (1.0,1.0) (2.0,2.0) (2.0,2.0) */ val accuracy = labelsAndPredictions.filter(p => p._1 == p._2).count()/testData.count() println("精准度:"+accuracy) //精准度:1 // Save and load model model.save(sc, "target/tmp/naiveBayesModel") val sameModel = NaiveBayesModel.load(sc, "target/tmp/naiveBayesModel") } }