TSception: Capturing Temporal Dynamics and Spatial Asymmetry from EEG for EmotionRecognition 学习笔记+源码
1.摘要
高时间分辨率和不对称的脑空间活跃性是脑电图(EEG)的基本属性,它是大脑中情感过程的基础。 为了学习脑电信号的动态时间和空间不对称性,从而实现准确和广义的情感识别,我们提出了一种多尺度卷积神经网络TSception。 TSception由动态的时间层、非对称的空间层和高级融合层组成,它们同时学习时间和通道维度上的特征表示。 动态时间层由多尺度一维卷积核组成,卷积核的长度与EEG的采样率有关,它学习EEG的动态时间和频率。 非对称空间层利用情绪的非对称脑电模式,学习区分性的全局和半脑表征。 学习到的空间表示将被一个高级融合层融合。 在DEAP和MAHNOB-HCI两个公开数据集上,使用更广义的交叉验证,对所提出的方法进行了评估。 将该网络的性能与SVM、KNN、FBFGMDM、FBTSC、无监督学习、DeepConvNet、ShallowConvNet、EEGNet等方法进行了比较。 在大多数实验中,TSCSeption比其他方法获得了更高的分类准确率和F1得分。https://github.com/yi-ding-cs/TSception
2.DEAP数据预处理
网络训练使用EAP和MAHNOB-HCI,以DEAP数据集为例,每个被试的数据集的结构为
删除每个被试实验前3秒的baseline, 然后将数据从512Hz向下采样到128Hz,然后用盲源分离方法去除眼电图(EOG)。 为了去除低频和高频噪声,对原始脑电信号进行4.0-45Hz的带通滤波,最后,将脑电通道平均到共同参考值。 每个维度的类标签从1到9,选择5作为阈值,将9个离散值投射到每个维度的低类和高类中。本研究仅使用唤醒和价维度。 由于深度神经网络具有更多的可训练参数,因此要对脑电中的情绪状态进行优化学习,需要大量的标记数据样本。 但是,如表1所列,在选定的数据集中,试验的数量很少。 为了克服这一挑战,通过将每个试验分成更小的、不重叠的4S片段来进行数据增强步骤(在训练部分加入这部分后面会提到)。 然后,这些片段被用来训练深度神经网络。
加载每个被试的数据,去除baseline
self.original_order = ['Fp1', 'AF3', 'F3', 'F7', 'FC5', 'FC1', 'C3', 'T7', 'CP5', 'CP1', 'P3', 'P7', 'PO3',
'O1', 'Oz', 'Pz', 'Fp2', 'AF4', 'Fz', 'F4', 'F8', 'FC6', 'FC2', 'Cz', 'C4', 'T8', 'CP6',
'CP2', 'P4', 'P8', 'PO4', 'O2'] ## 初始顺序
self.TS = ['Fp1', 'AF3', 'F3', 'F7', 'FC5', 'FC1', 'C3', 'T7', 'CP5', 'CP1', 'P3', 'P7', 'PO3','O1',
'Fp2', 'AF4', 'F4', 'F8', 'FC6', 'FC2', 'C4', 'T8', 'CP6', 'CP2', 'P4', 'P8', 'PO4', 'O2']
def load_data_per_subject(self, sub):
"""
This function loads the target subject's original file
Parameters
----------
sub: which subject to load
Returns
-------
data: (40, 32, 7680) label: (40, 4)
"""
sub += 1
if (sub < 10):
sub_code = str('s0' + str(sub) + '.dat')
else:
sub_code = str('s' + str(sub) + '.dat')
subject_path = os.path.join(self.data_path, sub_code)
subject = cPickle.load(open(subject_path, 'rb'), encoding='latin1')
label = subject['labels']
data = subject['data'][:, 0:32, 3 * 128:] # Excluding the first 3s of baseline
# data: 40 x 32 x 7680
# label: 40 x 4
# reorder the EEG channel to build the local-global graphs
# 这个作者的下一篇文章需要graph操作
data = self.reorder_channel(data=data, graph=self.graph_type)
print('data:' + str(data.shape) + ' label:' + str(label.shape))
return data, label
def reorder_channel(self, data, graph):
"""
This function reorder the channel according to different graph designs
Parameters
----------
data: (trial, channel, data)
graph: graph type
Returns
-------
reordered data: (trial, channel, data)
"""
if graph == 'TS':
graph_idx = self.TS # 去掉Fz、Cz、Pz、Oz的channnel
elif graph == 'O':
graph_idx = self.original_order # 正常channel
idx = []
for chan in graph_idx:
idx.append(self.original_order.index(chan))
return data[:, idx, :]
graph选择TS,data则去除Fz、Cz、Pz、Oz四个channnel,,最后的维度变为
![]()
标签分类
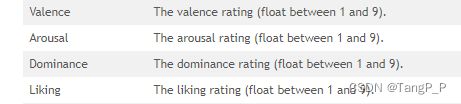
DEAP官网给出的标签类别
选取Valence与Arousal,设置阈值为5,大于5的label为1,小于等于5的label为0。
def label_selection(self, label):
"""
This function: 1. selects which dimension of labels to use
2. create binary label
Parameters
----------
label: (trial, 4)
Returns
-------
label: (trial,)
"""
if self.label_type == 'A':
label = label[:, 1]
elif self.label_type == 'V':
label = label[:, 0]
if self.args.num_class == 2:
label = np.where(label <= 5, 0, label)
label = np.where(label > 5, 1, label)
print('Binary label generated!')
return label数据分割成4S的片段 data格式 40*15*28*512 对应的label 40*15
def split(self, data, label, segment_length=4, overlap=0, sampling_rate=128):
"""
This function split one trial's data into shorter segments
Parameters
----------
data: (trial, channel, data)
data(40,28,7680)
label: (trial,)
data(40,)
segment_length: how long each segment is (e.g. 1s, 2s,...)
overlap: overlap rate
sampling_rate: sampling rate
Returns
-------
data:(trial, num_segment, channel, segment_legnth)
(40,15,28,512)
label:(trial, num_segment,)
(40,15)
"""
data_shape = data.shape # 40*28*7680
step = int(segment_length * sampling_rate * (1 - overlap))
data_segment = sampling_rate * segment_length # 分割后的信号采样点 128*4
data_split = []
number_segment = int((data_shape[2] - data_segment) // step)
for i in range(number_segment + 1):
data_split.append(data[:, :, (i * step):(i * step + data_segment)])
data_split_array = np.stack(data_split, axis=1)
label = np.stack([np.repeat(label[i], int(number_segment + 1)) for i in range(len(label))], axis=0)
print("The data and label are split: Data shape:" + str(data_split_array.shape) + " Label:" + str(
label.shape))
data = data_split_array
assert len(data) == len(label)
return data, label保存被试预处理后的数据集为 .hdf文件
def save(self, data, label, sub):
"""
This function save the processed data into target folder
Parameters
----------
data: the processed data
label: the corresponding label
sub: the subject ID
Returns
-------
None
"""
save_path = os.getcwd()
data_type = 'data_{}_{}_{}'.format(self.args.data_format, self.args.dataset, self.args.label_type)
save_path = os.path.join(save_path, data_type)
if not os.path.exists(save_path):
os.makedirs(save_path)
else:
pass
name = 'sub' + str(sub) + '.hdf'
save_path = os.path.join(save_path, name)
dataset = h5py.File(save_path, 'w')
dataset['data'] = data
dataset['label'] = label
dataset.close()完整的数据预处理过程,为了把data放入2D-CNN,data要多加一个维度,预处理后的数据集data的维度为40*15*1*28*512,label的维度为40*15
class PrepareData:
def __init__(self, args):
# init all the parameters here
# arg contains parameter settings
self.args = args
self.data = None
self.label = None
self.model = None
self.data_path = args.data_path
self.label_type = args.label_type
self.original_order = ['Fp1', 'AF3', 'F3', 'F7', 'FC5', 'FC1', 'C3', 'T7', 'CP5', 'CP1', 'P3', 'P7', 'PO3',
'O1', 'Oz', 'Pz', 'Fp2', 'AF4', 'Fz', 'F4', 'F8', 'FC6', 'FC2', 'Cz', 'C4', 'T8', 'CP6',
'CP2', 'P4', 'P8', 'PO4', 'O2'] ## 初始顺序
self.TS = ['Fp1', 'AF3', 'F3', 'F7', 'FC5', 'FC1', 'C3', 'T7', 'CP5', 'CP1', 'P3', 'P7', 'PO3','O1',
'Fp2', 'AF4', 'F4', 'F8', 'FC6', 'FC2', 'C4', 'T8', 'CP6', 'CP2', 'P4', 'P8', 'PO4', 'O2']
## 分成左右脑
self.graph_type = args.graph_type
def run(self, subject_list, split=False, expand=True, feature=False):
"""
Parameters
----------
subject_list: the subjects need to be processed
split: (bool) whether to split one trial's data into shorter segment
expand: (bool) whether to add an empty dimension for CNN
feature: (bool) whether to extract features or not
Returns
-------
The processed data will be saved './data___/sub0.hdf'
"""
for sub in subject_list:
data_, label_ = self.load_data_per_subject(sub)
# select label type here
label_ = self.label_selection(label_)
if split:
data_, label_ = self.split(
data=data_, label=label_, segment_length=self.args.segment,
overlap=self.args.overlap, sampling_rate=self.args.sampling_rate)
if expand:
# expand one dimension for deep learning(CNNs)
data_ = np.expand_dims(data_, axis=-3)
print('Data and label prepared for sub{}!'.format(sub))
print('data:' + str(data_.shape) + ' label:' + str(label_.shape))
print('----------------------')
self.save(data_, label_, sub) 3.网络结构
## TSception参数
args.model == 'TSception':
model = TSception(
num_classes=args.num_class, input_size=args.input_shape,
sampling_rate=args.sampling_rate, num_T=args.T, num_S=args.T,
hidden=args.hidden, dropout_rate=args.dropout)
num_class = 2
input_shape = (1,28,512)
samplig_rate = 128
T = 15
hidden = 32
dropout = 0.5
class TSception(nn.Module):
def conv_block(self, in_chan, out_chan, kernel, step, pool):
return nn.Sequential(
nn.Conv2d(in_channels=in_chan, out_channels=out_chan,
kernel_size=kernel, stride=step),
nn.LeakyReLU(),
nn.AvgPool2d(kernel_size=(1, pool), stride=(1, pool)))
def __init__(self, num_classes, input_size, sampling_rate, num_T, num_S, hidden, dropout_rate):
# input_size: 1 x EEG channel x datapoint
# sample_rate: 512
super(TSception, self).__init__()
self.inception_window = [0.5, 0.25, 0.125]
self.pool = 8
# by setting the convolutional kernel being (1,lenght) and the strids being 1 we can use conv2d to
# achieve the 1d convolution operation
self.Tception1 = self.conv_block(1, num_T, (1, int(self.inception_window[0] * sampling_rate)), 1, self.pool)
self.Tception2 = self.conv_block(1, num_T, (1, int(self.inception_window[1] * sampling_rate)), 1, self.pool)
self.Tception3 = self.conv_block(1, num_T, (1, int(self.inception_window[2] * sampling_rate)), 1, self.pool)
self.Sception1 = self.conv_block(num_T, num_S, (int(input_size[1]), 1), 1, int(self.pool*0.25))
self.Sception2 = self.conv_block(num_T, num_S, (int(input_size[1] * 0.5), 1), (int(input_size[1] * 0.5), 1),
int(self.pool*0.25))
self.fusion_layer = self.conv_block(num_S, num_S, (3, 1), 1, 4)
self.BN_t = nn.BatchNorm2d(num_T)
self.BN_s = nn.BatchNorm2d(num_S)
self.BN_fusion = nn.BatchNorm2d(num_S)
self.fc = nn.Sequential(
nn.Linear(num_S, hidden),
nn.ReLU(),
nn.Dropout(dropout_rate),
nn.Linear(hidden, num_classes)
)
def forward(self, x):
y = self.Tception1(x)
out = y
y = self.Tception2(x)
out = torch.cat((out, y), dim=-1) #最后一个维度拼接
y = self.Tception3(x)
out = torch.cat((out, y), dim=-1)
out = self.BN_t(out)
z = self.Sception1(out)
out_ = z
z = self.Sception2(out)
out_ = torch.cat((out_, z), dim=2)
out = self.BN_s(out_)
out = self.fusion_layer(out)
out = self.BN_fusion(out)
out = torch.squeeze(torch.mean(out, dim=-1), dim=-1)
out = self.fc(out)
return out
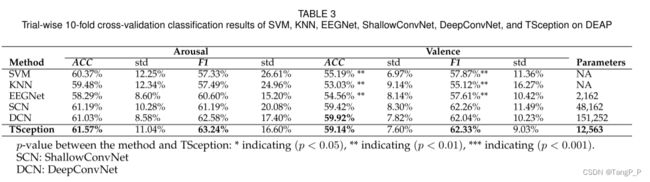
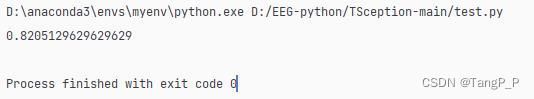
4.论文实验结果与复现
训练时间太长,训练了几个被试,其中被试1的训练数据,10折交叉的平均准确率为