Bayesian Model Averaging (BMA)的R实现
变量选择模型,贝叶斯模型平均法通过根据近似的后验模型概率对模型类中的最佳模型进行平均,说明了变量选择问题中固有的模型不确定性。
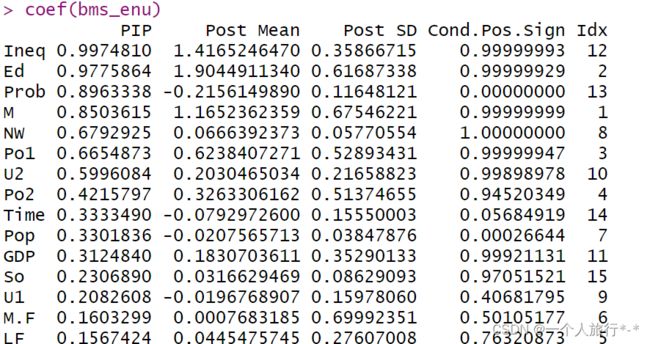
它对指定的生物标志物与结果的所有可能组合进行模型估计,并通过其后验模型概率对模型进行加权,同时调整协变量以确定给定变量对结果的影响程度。它产生后置包容概率(Posterior Inclusion Probabilities,PIP值),这是衡量每个变量相对于BMA模型中其他变量对结果的影响。
在R中有三个包可以实现BMA:BMA,BMS及BAS
实例:
set.seed(2011)
library(MASS)
data(UScrime); attach(UScrime)
UScrime1 <- (cbind(log(UScrime[,c(16,1,3:15)]), So))
noise<- matrix(rnorm(35*nrow(UScrime)), ncol=35)
colnames(noise)<- paste('noise', 1:35, sep='')
UScrime.log <- cbind(UScrime1,noise)
X <- UScrime1[,-1]; Y <- UScrime1[,1]
x <- UScrime.log[,-1]; y <- UScrime.log[,1]1.BMS
library(BMS)
bms_enu <- bms(UScrime1, mcmc="enumerate", g="UIP", mprior="uniform",
user.int=FALSE)
coef(bms_enu)2.BMA
library(BMA)
bma_enu <- iBMA.bicreg(X, Y, thresProbne0 = 5, verbose = TRUE, maxNvar = 30)
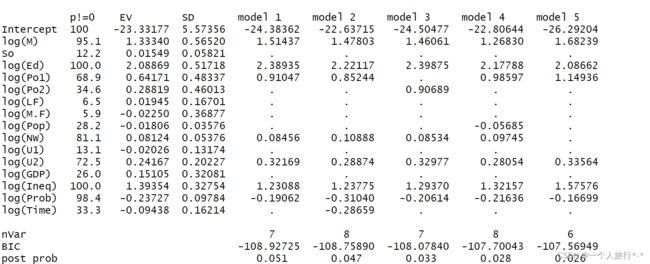
summary(bma_enu)library(MASS)
data(UScrime)
f <- formula(log(y) ~ log(M)+So+log(Ed)+log(Po1)+log(Po2)+log(LF)+
log(M.F)+ log(Pop)+log(NW)+log(U1)+log(U2)+
log(GDP)+log(Ineq)+log(Prob)+log(Time))
glm.out.crime <- bic.glm(f, data = UScrime, glm.family = gaussian())
summary(glm.out.crime)
3.BAS
library(BAS)
bas_enu<- bas.lm(y~., data=UScrime1, n.models=NULL, prior="ZS-null",
modelprior=uniform(), initprobs="Uniform")
coef(bas_enu)ref: Amini, S.M. and Parmeter, C.F., 2011. Bayesian model averaging in R. Journal of Economic and Social Measurement, 36(4), pp.253-287.
Berger, K., Eskenazi, B., Balmes, J., Kogut, K., Holland, N., Calafat, A.M. and Harley, K.G., 2019. Prenatal high molecular weight phthalates and bisphenol A, and childhood respiratory and allergic outcomes. Pediatric Allergy and Immunology, 30(1), pp.36-46.
bic.glm: Bayesian Model Averaging for generalized linear models. in BMA: Bayesian Model Averaging (rdrr.io)