【GNN笔记】GraphSAGE(四)
视频链接:【图神经网络】GNN从入门到精通
GNN中三种基础神经网络:GCN, GraphSAGE, GAT
文章目录
-
- 一、方法介绍
- 二、GraphSAGE伪代码
-
- ==聚合==
- ==采样==
- ==流程==
- ==补充要点==
- 三、GraphSAGE_minbatch伪代码
-
- 3.1 graph的全图变小图
- 3.2 采用小图进行信息聚合
- 四、GraphSAGE-Embedding
- 五、GraphSAGE的目录
- 六、dataCenter.py中`class DataCenter(object)`
- 七、models.py中的GraphSage
-
-
- `def __init__`
- `def forward`
- `aggregate`
- `def _get_unique_neighs_list` 函数
-
- 八、models.py中的Classification
- 九、utils.py中的`apply_model`
-
- 注释一:
- 附录:main.py文件
一、方法介绍
GraphSAGE:GraphSAGE (SAmple and aggreGatE)
GCN和GraphSAGE都是聚合节点的邻居信息获得下一层的节点表示.

二、GraphSAGE伪代码
- K K K :表示网络的层数
聚合
采样
假设采样样本大小=3 ,
R 1 ( v 1 ) = { v 2 , v 3 , v 4 , v 5 , v 6 } R_1(v_1)=\{v_2,v_3,v_4,v_5,v_6\} R1(v1)={v2,v3,v4,v5,v6},
R 1 ( v 2 ) = { v 1 , v 3 } R_1(v_2)=\{v_1,v_3\} R1(v2)={v1,v3}
v 1 v_1 v1的邻居数目超过3,直接采样抽取3个即可。
v 2 v_2 v2的邻居数目未超3,将其所有邻居取遍,然后再从 { v 1 , v 3 } \{v_1,v_3\} {v1,v3}中抽取1个。
- 采样最优
作者认为进行两层GraphSAGE(K=2)时,且 S 1 ⋅ S 2 < 500 S_1 \cdot S_2<500 S1⋅S2<500的时候采样效果最优。 S 1 S_1 S1表示在第一层采样的数目, S 2 S_2 S2表示在第二层采样的数目。
流程
当Agg(聚合)选择Mean函数时,邻居抽样的序列案例如下:

如图Fig.2-5所示graph,设采样的样本的大小sample_size=3
- v 1 v_1 v1的入度1-hop邻居有 N v 1 1 = { v 2 , v 3 , v 5 , v 6 } N^1_{v1}=\{v_2,v_3,v_5,v_6 \} Nv11={v2,v3,v5,v6},,采样的结果为 { v 6 , v 5 , v 2 } \{v_6,v_5,v_2\} {v6,v5,v2};
- v 2 v_2 v2的入度1-hop邻居有 N v 2 1 = { v 1 , v 3 } N^1_{v2}=\{v_1,v_3\} Nv21={v1,v3},采样的结果为${v_1,v_3,v_3}
补充要点
聚合函数
因为采样的节点不存在顺序性,所以聚合函数要求具有对称性。因此,作者给出了三个聚合函数:
三、GraphSAGE_minbatch伪代码
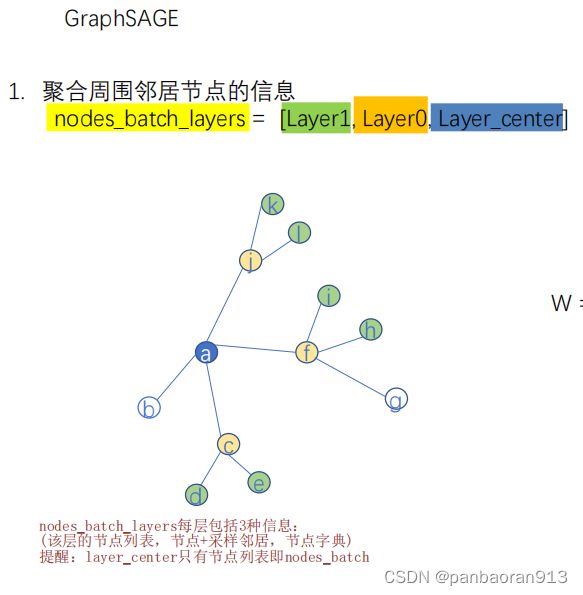
3.1 graph的全图变小图
step1:在B2层选择节点 a
step2:在B1层抽样3个邻居节点 c f j
step3:在B0层抽样2个邻居节点 d e, h i, l k
3.2 采用小图进行信息聚合
提醒:在进行信息聚合的时候输入的图结构是如Fig.3-3所示的,而非全部的图结构。
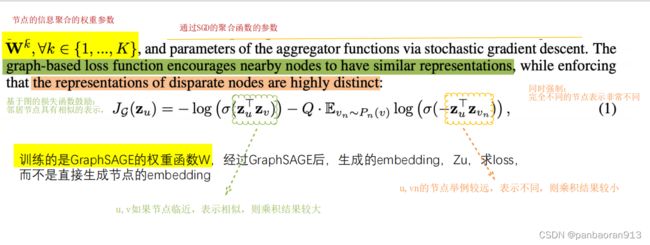
四、GraphSAGE-Embedding
提醒:loss fun训练的的权重参数,而不是embedding.
五、GraphSAGE的目录
- main的流程
六、dataCenter.py中class DataCenter(object)
class DataCenter(object):
"""docstring for DataCenter"""
def __init__(self, config):
super(DataCenter, self).__init__()
self.config = config
def load_dataSet(self, dataSet='cora'):
if dataSet == 'cora':
# 文件路径
cora_content_file = self.config['file_path.cora_content']
cora_cite_file = self.config['file_path.cora_cite']
#获得特征数据
feat_data = [] # 用于存放特征数据
labels = [] # label sequence of node 用于存放node的标签
node_map = {} # map node to Node_ID 字典
label_map = {} # map label to Label_ID 字典
with open(cora_content_file) as fp: # 打开数据集
for i,line in enumerate(fp): #对文件每行进行循环
info = line.strip().split() # 见注释一
feat_data.append([float(x) for x in info[1:-1]])
# info从id1到倒数第二个都是node的特征向量
node_map[info[0]] = i # node的编号,放入字典
if not info[-1] in label_map:
label_map[info[-1]] = len(label_map)
# node的分类数目<<<节点的个数的,如果该分类标签没有记录过,则放入字典
labels.append(label_map[info[-1]])# 用于存放node的标签
feat_data = np.asarray(feat_data)
labels = np.asarray(labels, dtype=np.int64)
# 获得adj矩阵
adj_lists = defaultdict(set)#设置默认字典,value为一个集合
with open(cora_cite_file) as fp:# 打开文件
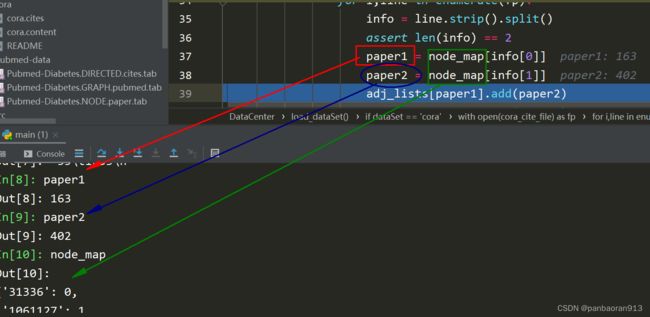
for i,line in enumerate(fp):# 对文件的每行进行循环
info = line.strip().split() # 间注释二
assert len(info) == 2 # 保证每行有2个数据,否则报错
paper1 = node_map[info[0]] # 将第一个节点的编号根据字典映射到int
paper2 = node_map[info[1]] # 将第二个节点的编号根据字典映射到int
adj_lists[paper1].add(paper2)# paper2作为paper1的邻居存入字典
adj_lists[paper2].add(paper1)#paper1作为paper2的邻居存入字典
assert len(feat_data) == len(labels) == len(adj_lists)
# 保证3者的数量相同
test_indexs, val_indexs, train_indexs = self._split_data(feat_data.shape[0])
# 分割test,val,train的idx
setattr(self, dataSet+'_test', test_indexs) # 见注释三
setattr(self, dataSet+'_val', val_indexs)
setattr(self, dataSet+'_train', train_indexs)
setattr(self, dataSet+'_feats', feat_data)
setattr(self, dataSet+'_labels', labels)
setattr(self, dataSet+'_adj_lists', adj_lists)
七、models.py中的GraphSage
def __init__
def forward
def forward中的运行
输入变量的形状
nb =[1,3,4,...] # 长度为2157 pre_neighs ={1:{1,344},3:{3, 197, 463, 601},{4, 170},...} 通过self.aggregate函数聚合节点的信息为aggregate_feats
center与第一层
aggregate
def aggregate(self, nodes, pre_hidden_embs, pre_neighs, num_sample=10):
""""
nodes: list,如图举例为[cfj]
pre_hidden_embs, [2708,1433]
pre_neighs: tuple,(层节点列表,节点+采样邻居ditc, 节点dict)
"""
unique_nodes_list, samp_neighs, unique_nodes = pre_neighs
assert len(nodes) == len(samp_neighs) # 确保每个节点都有对应的邻居采样
indicator = [(nodes[i] in samp_neighs[i]) for i in range(len(samp_neighs))]#如果节点在它自身的邻居节点范围内,返回True
assert (False not in indicator)# 确保所有的节点都在它自身的邻居范围内
if not self.gcn:# 如果不使用gcn算法
samp_neighs = [(samp_neighs[i] - set([nodes[i]])) for i in range(len(samp_neighs))]#只取邻居节点不要自身
# self.dc.logger.info('2')
# 对于节点特征[2708,1433],如果选择的节点=总节点,直接保留;否则选取节点特征
if len(pre_hidden_embs) == len(unique_nodes):
embed_matrix = pre_hidden_embs
else:
embed_matrix = pre_hidden_embs[torch.LongTensor(unique_nodes_list)]
# self.dc.logger.info('3')
# 构建相邻两层的节点的关系矩阵,比如[cfj]层和[dehilk]层的节点关系
mask = torch.zeros(len(samp_neighs), len(unique_nodes))# [2157,2558]
column_indices = [unique_nodes[n] for samp_neigh in samp_neighs for n in samp_neigh]
row_indices = [i for i in range(len(samp_neighs)) for j in range(len(samp_neighs[i]))]
mask[row_indices, column_indices] = 1
# self.dc.logger.info('4')
if self.agg_func == 'MEAN':
num_neigh = mask.sum(1, keepdim=True)#按行求和,保持和输入数据一个维度
mask = mask.div(num_neigh).to(embed_matrix.device)#归一化操作,除以按行求和的元素
aggregate_feats = mask.mm(embed_matrix)#[2157,1433]# 矩阵相乘,等价于聚合周围的邻居信息
elif self.agg_func == 'MAX':
# print(mask)
indexs = [x.nonzero() for x in mask == 1]
aggregate_feats = []
# self.dc.logger.info('5')
for feat in [embed_matrix[x.squeeze()] for x in indexs]:
if len(feat.size()) == 1:
aggregate_feats.append(feat.view(1, -1))
else:
aggregate_feats.append(torch.max(feat, 0)[0].view(1, -1))
aggregate_feats = torch.cat(aggregate_feats, 0)
# self.dc.logger.info('6')
return aggregate_feats
def _get_unique_neighs_list 函数
def _get_unique_neighs_list(self, nodes, num_sample=10):
# nodes =[1,3,4,...,] 共1024
_set = set
to_neighs = [self.adj_lists[int(node)] for node in nodes]
# 对邻居节点进行采样,如果大于邻居数据则进行采样
if not num_sample is None:
_sample = random.sample
samp_neighs = [_set(_sample(to_neigh, num_sample))
if len(to_neigh) >= num_sample else to_neigh for to_neigh in to_neighs]
else:
samp_neighs = to_neighs
# | 表示集合的并,将节点和采样后的邻居合并在一起
samp_neighs = [samp_neigh | set([nodes[i]]) for i, samp_neigh in enumerate(samp_neighs)]
# set.union()返回两个集合的并集,将所有(节点+采样邻居)的集合合并后变为list
_unique_nodes_list = list(set.union(*samp_neighs))
# 将所有节点和重新编译的编号设为dict
i = list(range(len(_unique_nodes_list)))
unique_nodes = dict(list(zip(_unique_nodes_list, i)))
# 返回 己点+采样后的邻居,节点和编号字典,所有的节点列表
return samp_neighs, unique_nodes, _unique_nodes_list
八、models.py中的Classification
- PyTorch的nn.Linear()详解
九、utils.py中的apply_model
注释一:
- torch.optim.SGD()
附录:main.py文件
import sys
import os
import torch
import argparse
import pyhocon
import random
from src.dataCenter import *
from src.utils import *
from src.models import *
parser = argparse.ArgumentParser(description='pytorch version of GraphSAGE')
parser.add_argument('--dataSet', type=str, default='cora')
parser.add_argument('--agg_func', type=str, default='MEAN')
parser.add_argument('--epochs', type=int, default=50)
parser.add_argument('--b_sz', type=int, default=20)
parser.add_argument('--seed', type=int, default=824)
parser.add_argument('--cuda', action='store_true',
help='use CUDA')
parser.add_argument('--gcn', action='store_true')
parser.add_argument('--learn_method', type=str, default='sup')
parser.add_argument('--unsup_loss', type=str, default='normal')
parser.add_argument('--max_vali_f1', type=float, default=0)
parser.add_argument('--name', type=str, default='debug')
parser.add_argument('--config', type=str, default='./src/experiments.conf')
args = parser.parse_args()
if torch.cuda.is_available():
if not args.cuda:
print("WARNING: You have a CUDA device, so you should probably run with --cuda")
else:
device_id = torch.cuda.current_device()
print('using device', device_id, torch.cuda.get_device_name(device_id))
device = torch.device("cuda" if args.cuda else "cpu")
print('DEVICE:', device)
if __name__ == '__main__':
random.seed(args.seed)
np.random.seed(args.seed)
torch.manual_seed(args.seed)
torch.cuda.manual_seed_all(args.seed)
# load config file
config = pyhocon.ConfigFactory.parse_file(args.config)
# load data
ds = args.dataSet
dataCenter = DataCenter(config)
dataCenter.load_dataSet(ds)
features = torch.FloatTensor(getattr(dataCenter, ds+'_feats')).to(device)
graphSage = GraphSage(config['setting.num_layers'], features.size(1), config['setting.hidden_emb_size'], features, getattr(dataCenter, ds+'_adj_lists'), device, gcn=args.gcn, agg_func=args.agg_func)
graphSage.to(device)
num_labels = len(set(getattr(dataCenter, ds+'_labels')))
classification = Classification(config['setting.hidden_emb_size'], num_labels)
classification.to(device)
unsupervised_loss = UnsupervisedLoss(getattr(dataCenter, ds+'_adj_lists'), getattr(dataCenter, ds+'_train'), device)
if args.learn_method == 'sup':
print('GraphSage with Supervised Learning')
elif args.learn_method == 'plus_unsup':
print('GraphSage with Supervised Learning plus Net Unsupervised Learning')
else:
print('GraphSage with Net Unsupervised Learning')
for epoch in range(args.epochs):
print('----------------------EPOCH %d-----------------------' % epoch)
graphSage, classification = apply_model(dataCenter, ds, graphSage, classification, unsupervised_loss, args.b_sz, args.unsup_loss, device, args.learn_method)
if (epoch+1) % 2 == 0 and args.learn_method == 'unsup':
classification, args.max_vali_f1 = train_classification(dataCenter, graphSage, classification, ds, device, args.max_vali_f1, args.name)
if args.learn_method != 'unsup':
args.max_vali_f1 = evaluate(dataCenter, ds, graphSage, classification, device, args.max_vali_f1, args.name, epoch)