【参赛记录】糖尿病遗传风险预测
【参赛记录】糖尿病遗传风险预测
- 一、选题背景
- 二、题目任务
-
- 2.1 数据集字段说明
- 2.2 训练集说明
- 2.3 测试集说明
- 三、评估指标
- 四、数据分析和处理
-
- 4.1 缺失值和异常值分析
-
- 缺失值
- 异常值
- 4.2 主成分分析
- 4.3 相关性分析
- 4.4 特征工程和数据预处理
- 五、模型构建与评估
-
- 5.1 随机森林
-
- Python实现
- 效果评估
- 5.2 高阶树模型
-
- 参数优化
- 模型训练和验证
- 效果评估
- 5.3 Stacking集成
-
- Stacking模型构建
- Python实现
- 模型评估
- 六、一些分析
一、选题背景
截至2022年,中国糖尿病患者近1.3亿。中国糖尿病患病原因受生活方式、老龄化、城市化、家族遗传等多种因素影响。同时,糖尿病患者趋向年轻化。
糖尿病可导致心血管、肾脏、脑血管并发症的发生。因此,准确诊断出患有糖尿病个体具有非常重要的临床意义。糖尿病早期遗传风险预测将有助于预防糖尿病的发生。
根据《中国2型糖尿病防治指南(2017年版)》,糖尿病的诊断标准是具有典型糖尿病症状(烦渴多饮、多尿、多食、不明原因的体重下降)且随机静脉血浆葡萄糖≥11.1mmol/L或空腹静脉血浆葡萄糖≥7.0mmol/L或口服葡萄糖耐量试验(OGTT)负荷后2h血浆葡萄糖≥11.1mmol/L。
在这次比赛中,您需要通过训练数据集构建糖尿病遗传风险预测模型,然后预测出测试数据集中个体是否患有糖尿病,和我们一起帮助糖尿病患者解决这“甜蜜的烦恼”。
二、题目任务
2.1 数据集字段说明
编号 :标识个体身份的数字;
性别:1表示男性,0表示女性;
出生年份:出生的年份;
体重指数:体重除以身高的平方,单位kg/m2;
糖尿病家族史:标识糖尿病的遗传特性,记录家族里面患有糖尿病的家属,分成三种标识,分别是父母有一方患有糖尿病、叔叔或者姑姑有一方患有糖尿病、无记录;
舒张压:心脏舒张时,动脉血管弹性回缩时,产生的压力称为舒张压,单位mmHg;
口服耐糖量测试:诊断糖尿病的一种实验室检查方法。比赛数据采用120分钟耐糖测试后的血糖值,单位mmol/L;
胰岛素释放实验:空腹时定量口服葡萄糖刺激胰岛β细胞释放胰岛素。比赛数据采用服糖后120分钟的血浆胰岛素水平,单位pmol/L;
肱三头肌皮褶厚度:在右上臂后面肩峰与鹰嘴连线的重点处,夹取与上肢长轴平行的皮褶,纵向测量,单位cm;
患有糖尿病标识:数据标签,1表示患有糖尿病,0表示未患有糖尿病。
2.2 训练集说明
训练集一共有5070条数据,用于构建预测模型。数据的字段有编号、性别、出生年份、体重指数、糖尿病家族史、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度、患有糖尿病标识(最后一列),您也可以通过特征工程技术构建新的特征。
2.3 测试集说明
测试集一共有1000条数据,用于验证预测模型的性能。数据的字段有编号、性别、出生年份、体重指数、糖尿病家族史、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度。
三、评估指标
对于提交的结果,系统会采用二分类任务中的F1-score指标进行评价,F1-score越大说明预测模型性能越好,F1-score的定义如下:
F 1 _ S c o r e = 2 1 1 p r e c i s i o n + 1 r e c a l l F1\_Score=2\frac{1}{\frac{1}{precision} + \frac{1}{recall}} F1_Score=2precision1+recall11
其中:
p r e c i s i o n = 预 测 并 且 实 际 患 有 糖 尿 病 的 个 体 数 量 预 测 患 有 糖 尿 病 的 个 体 数 量 precision=\frac{预测并且实际患有糖尿病的个体数量}{预测患有糖尿病的个体数量} precision=预测患有糖尿病的个体数量预测并且实际患有糖尿病的个体数量
r e c a l l = 预 测 且 实 际 患 有 糖 尿 病 的 个 体 数 量 实 际 患 有 糖 尿 病 的 个 体 数 量 recall=\frac{预测且实际患有糖尿病的个体数量}{实际患有糖尿病的个体数量} recall=实际患有糖尿病的个体数量预测且实际患有糖尿病的个体数量
四、数据分析和处理
4.1 缺失值和异常值分析
缺失值
| 特征 | 缺失比重 | 初步处理意见 |
|---|---|---|
| 舒张压 | 4.87% | 可以不处理 |
| 胰岛素释放实验 | 76.49% | 使用正常值范围内中数代替 |
异常值
| 特征 | 异常值 | 异常比重 | 初步处理意见 |
|---|---|---|---|
| 体重指数 | 异常为0 | 80.16% | 不处理 |
| 口服耐糖量测试 | 异常为-1/0 | 4.97% | 可以不处理 |
| 胰岛素释放实验 | 异常为0 | 68.60% | 使用正常值范围内中数代替 |
| 肱三头肌皮褶厚度 | 异常为0 | 67.69% | 使用正常值范围内中数代替 |
进一步分析发现,所有确实和异常数据在测试集和训练集中的比重相近,故目前本模型对所有缺失和异常数据采用不处理的方式。
4.2 主成分分析
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
from dataloader import Dataset
train_data, train_label = Dataset(is_train=True).get_dataset()
print(train_data)
print(train_label)
def PCA_(X):
# 标准化
X_std = StandardScaler().fit(X).transform(X)
# 构建协方差矩阵
cov_mat = np.cov(X_std.T)
# 特征值和特征向量
eigen_vals, eigen_vecs = np.linalg.eig(cov_mat)
# 求出特征值的和
tot = sum(eigen_vals)
# 求出每个特征值占的比例
var_exp = [(i / tot) for i in eigen_vals]
print(var_exp)
cum_var_exp = np.cumsum(var_exp)
# 绘图,展示各个属性贡献量
plt.bar(range(len(eigen_vals)), var_exp, width=1.0, bottom=0.0, alpha=0.5, label='individual explained variance')
plt.step(range(len(eigen_vals)), cum_var_exp, where='post', label='cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal components')
plt.legend(loc='best')
plt.savefig("pca.png")
var = train_data.values
PCA_(var)
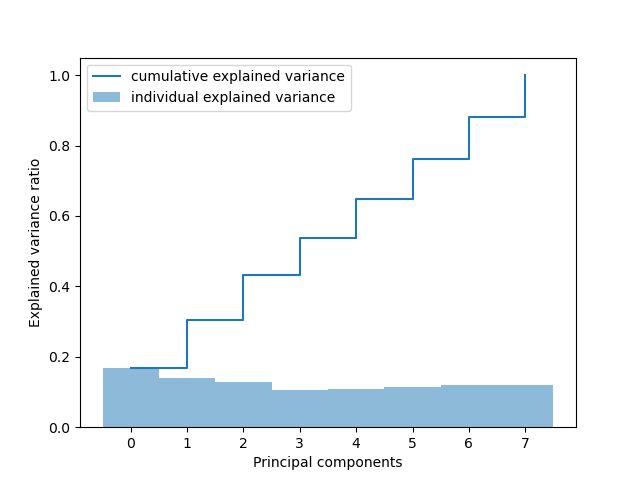
通过PCA分析发现,所有特征的贡献度相近,无极低或极高的特征,故不进行维度规约,所有特征直接参与模型构建。
4.3 相关性分析
import seaborn as sns
import matplotlib.pyplot as plt
from dataloader import Dataset
train_data, train_label = Dataset(is_train=True).get_dataset()
print(train_data)
print(train_label)
corr = train_data.corr()
print(corr)
plt.rcParams['font.sans-serif'] = ['Microsoft YaHei']
fig, ax = plt.subplots(figsize=(6, 6), facecolor='w')
# 指定颜色带的色系
sns.heatmap(train_data.corr(), annot=True, vmax=1, square=True, cmap="Blues", fmt='.2g')
plt.title('相关性热力图')
plt.show()
fig.savefig('./corr.png', bbox_inches='tight', transparent=True)
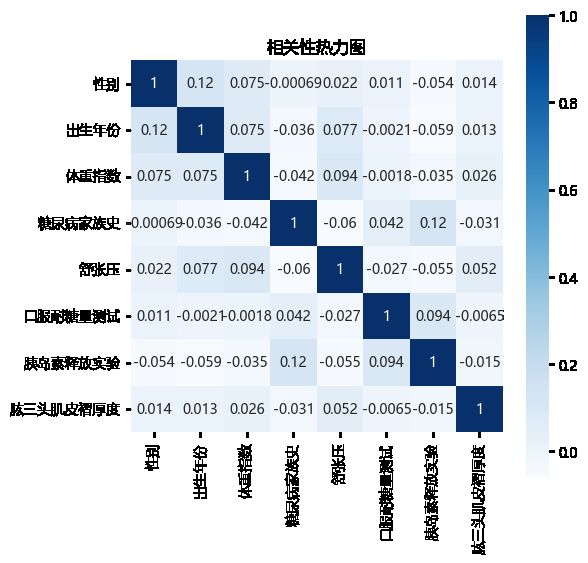
相关性热力图显示,不同特征之间没有明显的相关性(正相关或负相关),所以所有特征可以直接参与模型构建。
4.4 特征工程和数据预处理
import pandas as pd
class Dataset:
def __init__(self, is_train=True):
self.is_train = is_train
self.train_data = pd.DataFrame()
self.train_label = pd.DataFrame()
self.test_data = pd.DataFrame()
# 数据预处理
data = pd.read_csv('比赛训练集.csv', encoding='gbk')
data2 = pd.read_csv('比赛测试集.csv', encoding='gbk')
# 特征工程
data['出生年份'] = 2022 - data['出生年份'] # 换成年龄
data2['出生年份'] = 2022 - data2['出生年份'] # 换成年龄
# 糖尿病家族史
"""
无记录
叔叔或者姑姑有一方患有糖尿病/叔叔或姑姑有一方患有糖尿病
父母有一方患有糖尿病
"""
def FHOD(a):
if a == '无记录':
return 0
elif a == '叔叔或者姑姑有一方患有糖尿病' or a == '叔叔或姑姑有一方患有糖尿病':
return 1
else:
return 2
data['糖尿病家族史'] = data['糖尿病家族史'].apply(FHOD)
data['舒张压'] = data['舒张压'].fillna(-1)
data2['糖尿病家族史'] = data2['糖尿病家族史'].apply(FHOD)
data2['舒张压'] = data2['舒张压'].fillna(-1)
self.train_label = data['患有糖尿病标识']
self.train_data = data.drop(['编号', '患有糖尿病标识'], axis=1)
self.test_data = data2.drop(['编号'], axis=1)
def get_dataset(self):
if self.is_train:
return self.train_data, self.train_label
else:
return self.test_data
五、模型构建与评估
5.1 随机森林
随机森林是机器学习中十分常用的算法,也是bagging集成策略中最实用的算法之一。那么随机和森林分别是什么意思呢?森林应该比较好理解,分别建立了多个决策树,把它们放到一起不就是森林吗?这些决策树都是为了解决同一任务建立的,最终的目标也都是一致的,最后将其结果来平均即可。

Python实现
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import f1_score
from sklearn.model_selection import KFold, cross_val_score
from dataloader import Dataset
"""
Get the train set and validation set
"""
train_data, train_label = Dataset(is_train=True).get_dataset()
train_data = train_data.values
train_label = train_label.values
validation_data = Dataset(is_train=False).get_dataset()
validation_data = validation_data.values
def validation(model):
"""
Predict on validation set
:param model: trained model
:return: NULL
"""
pred = model.predict(validation_data)
pre_y = pd.DataFrame(pred)
pre_y['label'] = pre_y[0]
result = pd.read_csv('提交示例.csv')
result['label'] = pre_y['label']
result.to_csv('baseline.csv', index=False)
def kf_train(estimators):
"""
Train model using KFold for validation
:param estimators: n_estimators for RF model
:return: NUll
"""
clf = RandomForestClassifier(n_estimators=estimators, random_state=2022)
F1_score = 0
i = 1
kf = KFold(n_splits=5, shuffle=True)
for train_data_idx, test_data_idx in kf.split(train_data):
print("\n Fold: %d" % i)
train_data_ = train_data[train_data_idx]
train_label_ = train_label[train_data_idx]
test_data = train_data[test_data_idx]
test_label = train_label[test_data_idx]
i = i + 1
scores = cross_val_score(clf, train_data_, train_label_, cv=5, scoring='accuracy')
print("Accuracy: %f (+/- %f)" % (scores.mean(), scores.std()))
clf.fit(train_data_, train_label_)
print("Train Score: %f" % (clf.score(train_data_, train_label_)))
print("Test Score: %f" % (clf.score(test_data, test_label)))
pred = clf.predict(test_data)
f1 = f1_score(test_label, pred)
F1_score = F1_score + f1
print("F1 Score: %f" % f1)
print("AVG F1: %f" % (F1_score / 5))
def train(estimators):
"""
Train model
:param estimators: n_estimators for RF model
:return: Trained model
"""
clf = RandomForestClassifier(n_estimators=estimators, random_state=2022)
clf.fit(train_data, train_label)
return clf
def get_params():
"""
Applying KFold for optimize the main params for RF
:return: n_estimators for RF
"""
kf = KFold(n_splits=5, shuffle=True)
max_score_list = []
for train_data_idx, test_data_idx in kf.split(train_data):
tr_data = train_data[train_data_idx]
tr_label = train_label[train_data_idx]
te_data = train_data[test_data_idx]
te_label = train_label[test_data_idx]
score_lis = []
for ii in range(0, 200, 10):
rfc = RandomForestClassifier(n_estimators=ii + 1,
n_jobs=-1,
random_state=2022)
rfc.fit(tr_data, tr_label)
score = f1_score(te_label, rfc.predict(te_data))
score_lis.append(score)
max_score_list.append((score_lis.index(max(score_lis)) * 10) + 1)
print("Max F1_Score: %.4f, N = %d" % (max(score_lis), (score_lis.index(max(score_lis)) * 10) + 1))
plt.figure(figsize=[20, 5])
plt.plot(range(1, 201, 10), score_lis)
plt.show()
return np.mean(np.array(max_score_list))
if __name__ == "__main__":
n = get_params()
print(n)
kf_train(estimators=int(n))
classifier = train(estimators=int(n))
validation(model=classifier)
效果评估
由于没有标注的测试集,这里使用KFold验证方法,求所有Fold的F1得分平均值作为评估依据。效果如下:
KFold 验证:AVG F1: 0.944644
验证集得分:0.96594
5.2 高阶树模型
使用LightGBM模型进行训练和预测。
参数优化
import pandas as pd
import lightgbm as lgb
from dataloader import Dataset
train_data, train_label = Dataset(is_train=True).get_dataset()
print(train_data)
print(train_label)
# 数据转换
print('数据转换')
lgb_train = lgb.Dataset(train_data, train_label, free_raw_data=False)
# 设置初始参数--不含交叉验证参数
print('设置参数')
params = {
'boosting_type': 'gbdt',
'objective': 'binary',
'metric': 'auc',
'nthread': 4,
'learning_rate': 0.05
}
# 交叉验证(调参)
print('交叉验证')
max_auc = float('0')
best_params = {}
# 准确率
print("调参1:提高准确率")
for num_leaves in range(5, 100, 5):
for max_depth in range(3, 8, 1):
params['num_leaves'] = num_leaves
params['max_depth'] = max_depth
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['num_leaves'] = num_leaves
best_params['max_depth'] = max_depth
if 'num_leaves' and 'max_depth' in best_params.keys():
params['num_leaves'] = best_params['num_leaves']
params['max_depth'] = best_params['max_depth']
# 过拟合
print("调参2:降低过拟合")
for max_bin in range(5, 256, 10):
for min_data_in_leaf in range(1, 102, 10):
params['max_bin'] = max_bin
params['min_data_in_leaf'] = min_data_in_leaf
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['max_bin'] = max_bin
best_params['min_data_in_leaf'] = min_data_in_leaf
if 'max_bin' and 'min_data_in_leaf' in best_params.keys():
params['min_data_in_leaf'] = best_params['min_data_in_leaf']
params['max_bin'] = best_params['max_bin']
print("调参3:降低过拟合")
for feature_fraction in [0.6, 0.7, 0.8, 0.9, 1.0]:
for bagging_fraction in [0.6, 0.7, 0.8, 0.9, 1.0]:
for bagging_freq in range(0, 50, 5):
params['feature_fraction'] = feature_fraction
params['bagging_fraction'] = bagging_fraction
params['bagging_freq'] = bagging_freq
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['feature_fraction'] = feature_fraction
best_params['bagging_fraction'] = bagging_fraction
best_params['bagging_freq'] = bagging_freq
if 'feature_fraction' and 'bagging_fraction' and 'bagging_freq' in best_params.keys():
params['feature_fraction'] = best_params['feature_fraction']
params['bagging_fraction'] = best_params['bagging_fraction']
params['bagging_freq'] = best_params['bagging_freq']
print("调参4:降低过拟合")
for lambda_l1 in [1e-5, 1e-3, 1e-1, 0.0, 0.1, 0.3, 0.5, 0.7, 0.9, 1.0]:
for lambda_l2 in [1e-5, 1e-3, 1e-1, 0.0, 0.1, 0.4, 0.6, 0.7, 0.9, 1.0]:
params['lambda_l1'] = lambda_l1
params['lambda_l2'] = lambda_l2
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['lambda_l1'] = lambda_l1
best_params['lambda_l2'] = lambda_l2
if 'lambda_l1' and 'lambda_l2' in best_params.keys():
params['lambda_l1'] = best_params['lambda_l1']
params['lambda_l2'] = best_params['lambda_l2']
print("调参5:降低过拟合2")
for min_split_gain in [0.0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0]:
params['min_split_gain'] = min_split_gain
cv_results = lgb.cv(
params,
lgb_train,
seed=1,
nfold=5,
metrics=['auc'],
early_stopping_rounds=10,
verbose_eval=True
)
mean_auc = pd.Series(cv_results['auc-mean']).max()
boost_rounds = pd.Series(cv_results['auc-mean']).idxmax()
if mean_auc >= max_auc:
max_auc = mean_auc
best_params['min_split_gain'] = min_split_gain
if 'min_split_gain' in best_params.keys():
params['min_split_gain'] = best_params['min_split_gain']
print(best_params)
优化结果:
{'num_leaves': 95,
'max_depth': 7,
'max_bin': 255,
'min_data_in_leaf': 11,
'feature_fraction': 0.8,
'bagging_fraction': 0.9,
'bagging_freq': 30,
'lambda_l1': 1e-05,
'lambda_l2': 1e-05,
'min_split_gain': 0.0}
模型训练和验证
import pandas as pd
import lightgbm
from dataloader import Dataset
from sklearn.model_selection import train_test_split, KFold, cross_val_score
from sklearn.metrics import f1_score
clf = lightgbm.LGBMClassifier(
boosting_type='gbdt',
objective='binary',
learning_rate=0.05,
metric='auc',
num_leaves=95,
max_depth=7,
max_bin=255,
min_data_in_leaf=11,
feature_fraction=0.8,
bagging_fraction=0.9,
bagging_freq=30,
lambda_l1=1e-05,
lambda_l2=1e-05,
min_split_gain=0.0
)
"""
Get the train set and validation set
"""
train_data, train_label = Dataset(is_train=True).get_dataset()
train_data = train_data.values
train_label = train_label.values
validation_data = Dataset(is_train=False).get_dataset()
validation_data = validation_data.values
def validation(model):
"""
Predict on validation set
:param model: trained model
:return: NULL
"""
pred = model.predict(validation_data)
pre_y = pd.DataFrame(pred)
pre_y['label'] = pre_y[0]
result = pd.read_csv('提交示例.csv')
result['label'] = pre_y['label']
result.to_csv('baseline.csv', index=False)
def kf_train():
"""
Train model using KFold for validation
:return: NUll
"""
F1_score = 0
i = 1
kf = KFold(n_splits=5, shuffle=True)
for train_data_idx, test_data_idx in kf.split(train_data):
print("\n Fold: %d" % i)
train_data_ = train_data[train_data_idx]
train_label_ = train_label[train_data_idx]
test_data = train_data[test_data_idx]
test_label = train_label[test_data_idx]
i = i + 1
scores = cross_val_score(clf, train_data_, train_label_, cv=5, scoring='accuracy')
print("Accuracy: %f (+/- %f)" % (scores.mean(), scores.std()))
clf.fit(train_data_, train_label_)
print("Train Score: %f" % (clf.score(train_data_, train_label_)))
print("Test Score: %f" % (clf.score(test_data, test_label)))
pred = clf.predict(test_data)
f1 = f1_score(test_label, pred)
F1_score = F1_score + f1
print("F1 Score: %f" % f1)
print("AVG F1: %f" % (F1_score / 5))
def train():
"""
Train model
:return: Trained model
"""
clf.fit(train_data, train_label)
return clf
if __name__ == "__main__":
kf_train()
model = train()
validation(model=model)
效果评估
KFold 验证得分:0.940159
验证集得分:0.95449
5.3 Stacking集成
Stacking方法也是经典的集成学习方法,其基础思想与Bagging类似,都是结合多个模型的输出来完成最后的预测。但两者有以下不同:
- Bagging是采取投票或平均的方式来处理N个基模型的输出,Stacking方法是训练一个模型用于组合之前的基模型。
- Bagging中每个基模型的训练集是通过bootstrap抽样得到的,不尽相同。而Stacking方法中每个模型的训练集是一样的,使用全部的训练集来训练。 使用Stacking方法时,常常采用交叉验证的方法来训练基模型。

Stacking模型构建
Python实现
import pandas as pd
from sklearn.metrics import f1_score
from sklearn.model_selection import cross_val_score, KFold
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from lightgbm import LGBMClassifier
from xgboost import XGBClassifier
from mlxtend.classifier import StackingCVClassifier
from dataloader import Dataset
"""
Get train data and validation data
"""
train_data, train_label = Dataset(is_train=True).get_dataset()
train_data = train_data.values
train_label = train_label.values
validation_data = Dataset(is_train=False).get_dataset()
validation_data = validation_data.values
def validation(pred):
"""
Perform prediction on validation set
:param pred: prediction generated by model
:return: NULL
"""
pre_y = pd.DataFrame(pred)
pre_y['label'] = pre_y[0]
result = pd.read_csv('提交示例.csv')
result['label'] = pre_y['label']
result.to_csv('baseline.csv', index=False)
def test(pred, test_label):
"""
Perform test on split test data
:param pred: prediction generated by model
:param test_label: target label
:return: F1 Score on test dataset
"""
correct = 0
for i in range(len(pred)):
if pred[i] == test_label[i]:
correct = correct + 1
print("Correct : %f" % (correct / len(pred)))
f1 = f1_score(y_true=test_label, y_pred=pred)
print("F1_Score: %f" % f1)
return f1
"""
Construct model
"""
RANDOM_SEED = 42
clf1 = KNeighborsClassifier(n_neighbors=5)
clf2 = RandomForestClassifier(random_state=RANDOM_SEED)
clf3 = GaussianNB()
clf4 = SVC(gamma='auto')
clf5 = LogisticRegression(random_state=0, max_iter=180)
clf6 = XGBClassifier(learning_rate=0.05, objective='binary:logitraw', eval_metric='auc', use_label_encoder=False)
clf7 = LGBMClassifier(boosting_type='gbdt',
objective='binary',
learning_rate=0.05,
metric='auc',
num_leaves=95,
max_depth=7,
max_bin=255,
min_data_in_leaf=11,
feature_fraction=0.8,
bagging_fraction=0.9,
bagging_freq=30,
lambda_l1=1e-05,
lambda_l2=1e-05,
min_split_gain=0.0)
meta_clf = LogisticRegression(random_state=0, max_iter=180)
s_clf = StackingCVClassifier(classifiers=[clf1, clf2, clf3, clf4, clf5, clf5, clf7],
meta_classifier=meta_clf,
random_state=RANDOM_SEED)
def train(tr_data, tr_label, te_data, te_label):
"""
Perform CV validation on each base learner
:param tr_data: train data
:param tr_label: train label
:param te_data: test data
:param te_label: test label
:return: NULL
"""
for clf, label in zip([clf1, clf2, clf3, clf4, clf5, clf6, clf7, s_clf],
['KNN', 'Random Forest', 'Naive Bayes', 'SVM', 'LR', 'XGB', 'LGB', 'Stacking CLF']):
scores = cross_val_score(clf, tr_data, tr_label, cv=5, scoring='accuracy')
print("\nAccuracy: %f (+/- %f) [%s]" % (scores.mean(), scores.std(), label))
clf.fit(tr_data, tr_label)
print("Train Score: %f [%s]" % (clf.score(tr_data, tr_label), label))
print("Test Score: %f [%s]" % (clf.score(te_data, te_label), label))
s_clf.fit(train_data, train_label)
print("\nTRAIN SCORE: %f" % s_clf.score(train_data, train_label))
def main():
"""
Main function. Perform train and validation
:return: NULL
"""
F1_score = 0
i = 1
kf = KFold(n_splits=5, shuffle=True)
for train_data_idx, test_data_idx in kf.split(train_data):
print("\n%d" % i)
i = i + 1
train_data_ = train_data[train_data_idx]
train_label_ = train_label[train_data_idx]
test_data = train_data[test_data_idx]
test_label = train_label[test_data_idx]
train(train_data_, train_label_, test_data, test_label)
print("TEST SCORE: %f" % s_clf.score(test_data, test_label))
prediction = s_clf.predict(test_data)
F1_score = F1_score + test(prediction, test_label)
print("\nAVG_F1: %f" % (F1_score / 5))
s_clf.fit(train_data, train_label)
prediction = s_clf.predict(validation_data)
validation(prediction)
if __name__ == "__main__":
main()
模型评估
KFold 验证得分:0.995050
验证集得分:0.95951
六、一些分析
起初,笔者想训练一个多层感知机(MLP)进行二分类任务,但是随后发现多层感知机的性能甚至远不如简单的逻辑回归和支持向量机。这可能是由于数据过少和维度较少导致的。
从结果来看,随机森林的效果是最好的,高阶树模型(LightGBM)的性能最差。本被笔者给予厚望的Stacking集成模型,性能也不如随机森林。这可能是由于Stacking模型中各基分类器没有进行参数优化导致的。
此外,文中任何一个模型都直接使用原始数据,没有对数据进行进一步的处理,某些特征有较多的缺失值可能会影响性能。
