Randomization Tests
Randomization Tests
Constrained ordination methods such as CCA and RDA find the ' best possible' relationship (defined in a mathematical sense) between species composition and the environment. Therefore, if we correlate sample scores with environmental variables, and peform conventional statistical tests, we are almost guaranteed to get a 'significant' result, even with random data. So we are interested in determining whether or observed ordination is 'stronger', or the species-environment relationship is higher, than expected due to chance.
In eigenanalysis-based methods, the eigenvalue is a reasonable measure of the strength of an ordination axis. The sum of all constrained eigenvalues (or 'trace') is a reasonable measure of the strength of the ordination. But unfortunately, there is easy connection betwen these numbers and a standard statistical distribution such as the Normal, Poisson, Chi-squared, and the like. So classical statistical testing is problematic.
Fortunately, it is possible to let the data themselves define the statistical distribution of eigenvalues expected under the null hypothesis. Such a method is called a permutation test, or Monte Carlo Permutation Procedure (MCPP). Permutation tests are special cases of randomization tests, i.e. tests that use randomly generated numbers for statistical inference.
The availability of fast computers has made permutation tests increasingly feasible, even for large data sets. Since such methods require no particular assumptions concerning statistical distributions (with the exception of the important assumption of independent observations), permutation tests are increasingly applied even in the context of traditional statistical tests (e.g. correlation, t-tests, ANOVAS, etc.).
The procedure for a randomization test is:
1.Devise a test statistic which is large if your hypothesized process is strong, and small if it is weak (you could
do it the other way around, but let us ignore this for now).
2.Define your null hypothesis.
3.Create a new data set consisting of your data, randomly rearranged. Exactly how it is rearranged depends on
your null hypothesis.
4.Calculate your test statistic for this data set, and compare it to your true value.
5.Repeat steps 3 and 4 many times (preferably several hundred).
6.If your true test statistic is greater than 95% of the random values, then you can reject the null hypothesis at
p<0.05. (be careful about whether you are performing a one tailed vs. two tailed test - if the latter, you will
need to use a 97.5% cutoff).
This method may seem somewhat magical, or even circular - how can you get any information out of randomness?
It is because you are answering the question directly: "How likely is it that if the null hypothesis were true, I would
observe a value this extreme just due to chance?" It is worth knowing that Fisher used randomization tests to test the value of the t-test, F-tests, etc. Many people are now promoting the use of randomization tests even when
parametric and nonparametric tests exist. Statistical Educators are beginning to use randomization tests as the
introduction to statistics, because in many ways it is easier to grasp. See Manly (1992) for more information about
randomization tests in ecology.
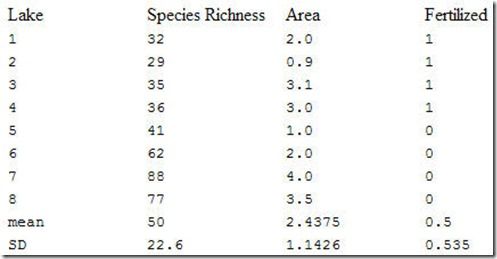
Let us take the previously given hypothetical example (in Statistics and in Multiple Regression) of invertebrate species richness in lakes.
Let us assume we are interested in testing whether the relationship between richness and area is stronger than expected due to chance. A reasonable measure of the strength of the relationship is r2. In this case, r2=0.4258. What value of r2 would we expect if the null hypothesis were true? In other words, what value would we expect if the relationship between richness and area were random? It would be the value we would get if the richnesses we observed were randomly assigned to the areas. But for different random assignments, we would get different values for r2. So the key to permutation tests is to determine the proportion of the random r2s that are greater than our observed r2. If very few (conventionally, we choose less than 5%, or alpha = 0.05) of our random values are higher than our observed ones, we reject the null hypothesis.
The following are a set of r2 calculated from 42 random permutations of richness and area:
0.5950 0.0894 0.0259 0.0047 0.2879 0.1649
0.0068 0.4786 0.0842 0.0066 0.0635 0.1839
0.0493 0.4901 0.1810 0.0001 0.3674 0.1496
0.0501 0.0434 0.0544 0.0166 0.0028 0.0838
0.0016 0.4809 0.0072 0.0643 0.0107 0.0101
0.3152 0.0015 0.0315 0.0094 0.0084 0.0000
0.0807 0.1322 0.3450 0.0004 0.1151 0.0125
Note that four of these values (0.4786, 0.4809, 0.4901, 0.5950) are greater than our true value of 0.4258. This means that p = 4/42 = 0.09525. If we choose the conventional cutoff of p=0.05, we fail to reject the null hypothesis that richness is unrelated to area. However, since the p value is close to the cutoff, we should be aware that a moderately increased sample size might have resulted in a significant relationship. An increased number of iterations (i.e. random sequences) would give us a better estimate of p, but it would not increase the likelihood of significance.
It might be argued that in this data set, we have two very different kinds of lake: those that have been fertilized and those that have not. It is possible that the fertilization effect is so strong, that it is hard to detect an area effect. A better permutation test might be to define two permutation blocks: one would be the fertilized lakes, and the other would be the unfertilized lakes. The permutation test would be identical to the preceding example, except one would only reshuffle observations within fertilized lakes, and separately within unfertilized lakes. Such a test would be more likely to reveal subtle effects of area. If we reject the null hypothesis, we would conclude that area had an important effect within treatment groups.
Permutation tests in constrained ordination are in principle no different from the above. Instead of randomly reassigning x's to y's, we are randomly reassigning our environmental variables to species composition. All of the species in a sample will be reassigned to the environmental variables of another sample. Permutation blocks are fairly simple to define, especially in the context of partial ordination.
This article was created by Michael Palmer. http://ordination.okstate.edu/permute.htm