Python异常值检测——案例分析
目录
1.单个变量异常值检测
2. 双变量关系中的异常值检测
3. 使用线性回归来确定具有重大影响的数据点
4. 使用k最近邻算法找到离群值
5. 使用隔离森林算法查找异常
1.单个变量异常值检测
如果某个值离平均值有多个标准偏差,并且远离近似标准正态分布的值,则我们更倾向于将将其视为异常值。
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
import scipy.stats as scistat
data=pd.read_csv("F:\\python数据清洗\\04第四章\\covidtotals.csv")data.columnsIndex(['iso_code', 'lastdate', 'location', 'total_cases', 'total_deaths',
'total_cases_pm', 'total_deaths_pm', 'population', 'pop_density',
'median_age', 'gdp_per_capita', 'hosp_beds'],
dtype='object')
data.set_index('iso_code',inplace=True)
totvars=['location', 'total_cases', 'total_deaths', 'total_cases_pm', 'total_deaths_pm']
demovars=['population', 'pop_density', 'median_age', 'gdp_per_capita', 'hosp_beds']1.1 描述性统计
data_b=data.loc[:,totvars]
data_b.describe()| total_cases | total_deaths | total_cases_pm | total_deaths_pm | |
|---|---|---|---|---|
| count | 2.100000e+02 | 210.000000 | 210.000000 | 210.000000 |
| mean | 2.921614e+04 | 1770.714286 | 1355.357943 | 55.659129 |
| std | 1.363978e+05 | 8705.565857 | 2625.277497 | 144.785816 |
| min | 0.000000e+00 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 1.757500e+02 | 4.000000 | 92.541500 | 0.884750 |
| 50% | 1.242500e+03 | 25.500000 | 280.928500 | 6.154000 |
| 75% | 1.011700e+04 | 241.250000 | 1801.394750 | 31.777250 |
| max | 1.790191e+06 | 104383.000000 | 19771.348000 | 1237.551000 |
1.2 显示百分位数数据
data_b.quantile(np.arange(0.0,1.1,0.1))| total_cases | total_deaths | total_cases_pm | total_deaths_pm | |
|---|---|---|---|---|
| 0.0 | 0.0 | 0.0 | 0.0000 | 0.0000 |
| 0.1 | 22.9 | 0.0 | 17.9986 | 0.0000 |
| 0.2 | 105.2 | 2.0 | 56.2910 | 0.3752 |
| 0.3 | 302.0 | 6.7 | 115.4341 | 1.7183 |
| 0.4 | 762.0 | 12.0 | 213.9734 | 3.9566 |
| 0.5 | 1242.5 | 25.5 | 280.9285 | 6.1540 |
| 0.6 | 2514.6 | 54.6 | 543.9562 | 12.2452 |
| 0.7 | 6959.8 | 137.2 | 1071.2442 | 25.9459 |
| 0.8 | 16847.2 | 323.2 | 2206.2982 | 49.9658 |
| 0.9 | 46513.1 | 1616.9 | 3765.1363 | 138.9045 |
| 1.0 | 1790191.0 | 104383.0 | 19771.3480 | 1237.5510 |
1.3 偏度(skewness)&峰度(kurtosis)
偏度和峰度分别描述了分布的对称性和分布的尾部有多肥。如果变量以正态分布,则这两个指标都大大高于预期。
# 偏度skewness
data_b.skew()
----------------------------
total_cases 10.804275
total_deaths 8.929816
total_cases_pm 4.396091
total_deaths_pm 4.674417
dtype: float64
# 峰度kurtosis
data_b.kurtosis()
----------------------------
total_cases 134.979577
total_deaths 95.737841
total_cases_pm 25.242790
total_deaths_pm 27.238232
dtype: float641.4 测试数据的正态分布性
使用scipy库中的Shapiro-Wilk测试,输出测试的p值。正态分布的null假设可以在低于0.05的任何p值和95%的置信水平上被拒绝。
def testnorm(var,df):
stat,p=scistat.shapiro(df[var])
return p
testnorm("total_cases",data_b) #3.753789128593843e-29
testnorm("total_deaths",data_b) #4.3427896631016077e-29
testnorm("total_cases_pm",data_b) #1.3972683006509067e-23
testnorm("total_deaths_pm",data_b) #1.361060423265974e-25结果表明变量的分布在高显著水平下不是正态的。
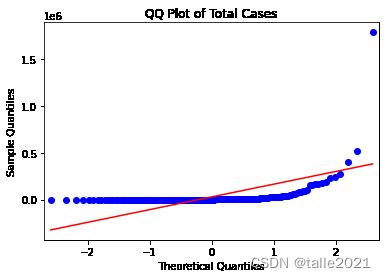
# 总病例数的分位数图(qq图),直线显示的是正态分布应有的外观
sm.qqplot(data_b[["total_cases"]].sort_values(['total_cases']),line='s')
plt.title('QQ Plot of Total Cases')
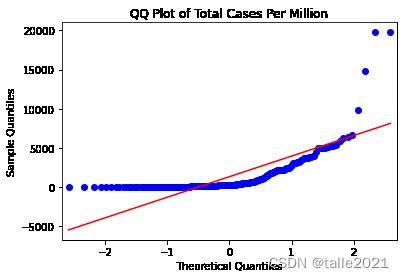
plt.show()# 每百万人口的总病例数的分位数图(qq图)
sm.qqplot(data_b[["total_cases_pm"]].sort_values(['total_cases_pm']),line='s')
plt.title('QQ Plot of Total Cases Per Million')
plt.show()qqplot的结果也表明,total_cases(总病例数)和total_cases_pm(每百万人口的总病例数)的分布都与正态分布有显著差异(红色直线显示的是正态分布应有的外观)
1.5 显示离群值范围
定义变量离群值的一种方法是按四分位数的距离计算。第三个四分位数(0.75)被称为上四分位数,第一个四分位数(0.25)被称为下四分位数,上四分位数和下四分位数之间的距离称为四分位距,也就是0.25~0.75分位数的范围。如果某个值离均值大于1.5倍,则认为该值是离群值。
def getoutliers():
dfout=pd.DataFrame(columns=data.columns,data=None)
for col in data_b.columns[1:]:
thirdq,firstq=data_b[col].quantile(0.75),data_b[col].quantile(0.25) #计算四分位数
range_q=1.5*(thirdq-firstq) #四分位距
high,low=range_q+thirdq,firstq-range_q #离群阈值
df=data.loc[(data[col]>high) | (data[col]| lastdate | location | total_cases | total_deaths | total_cases_pm | total_deaths_pm | population | pop_density | median_age | gdp_per_capita | hosp_beds | varname | threshlow | threshhigh | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BGD | 2020-06-01 | Bangladesh | 47153 | 650 | 286.315 | 3.947 | 164689383.0 | 1265.036 | 27.5 | 3523.984 | 0.80 | total_cases | -14736.125 | 25028.875 |
| BLR | 2020-06-01 | Belarus | 42556 | 235 | 4503.604 | 24.870 | 9449321.0 | 46.858 | 40.3 | 17167.967 | 11.00 | total_cases | -14736.125 | 25028.875 |
| BEL | 2020-06-01 | Belgium | 58381 | 9467 | 5037.354 | 816.852 | 11589616.0 | 375.564 | 41.8 | 42658.576 | 5.64 | total_cases | -14736.125 | 25028.875 |
| BRA | 2020-06-01 | Brazil | 514849 | 29314 | 2422.142 | 137.910 | 212559409.0 | 25.040 | 33.5 | 14103.452 | 2.20 | total_cases | -14736.125 | 25028.875 |
| CAN | 2020-06-01 | Canada | 90936 | 7295 | 2409.401 | 193.285 | 37742157.0 | 4.037 | 41.4 | 44017.591 | 2.50 | total_cases | -14736.125 | 25028.875 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ESP | 2020-05-31 | Spain | 239429 | 27127 | 5120.952 | 580.197 | 46754783.0 | 93.105 | 45.5 | 34272.360 | 2.97 | total_deaths_pm | -45.454 | 78.116 |
| SWE | 2020-06-01 | Sweden | 37542 | 4395 | 3717.298 | 435.180 | 10099270.0 | 24.718 | 41.0 | 46949.283 | 2.22 | total_deaths_pm | -45.454 | 78.116 |
| CHE | 2020-06-01 | Switzerland | 30779 | 1656 | 3556.367 | 191.343 | 8654618.0 | 214.243 | 43.1 | 57410.166 | 4.53 | total_deaths_pm | -45.454 | 78.116 |
| GBR | 2020-06-01 | United Kingdom | 274762 | 38489 | 4047.403 | 566.965 | 67886004.0 | 272.898 | 40.8 | 39753.244 | 2.54 | total_deaths_pm | -45.454 | 78.116 |
| USA | 2020-06-01 | United States | 1790191 | 104383 | 5408.389 | 315.354 | 331002647.0 | 35.608 | 38.3 | 54225.446 | 2.77 | total_deaths_pm | -45.454 | 78.116 |
outliers.varname.value_counts()total_deaths 36 total_cases 33 total_deaths_pm 28 total_cases_pm 17 Name: varname, dtype: int64
2. 双变量关系中的异常值检测
当某个值的分布与平均值没有明显偏离时,即使它不是极值,它也可能是异常值。当第二个变量具有某些值时,第一个变量的某些值就可能是异常值。(以下使用的还是之前的数据)
2.1 生成相关性矩阵
data.corr(method="pearson")| total_cases | total_deaths | total_cases_pm | total_deaths_pm | population | pop_density | median_age | gdp_per_capita | hosp_beds | |
|---|---|---|---|---|---|---|---|---|---|
| total_cases | 1.000000 | 0.932079 | 0.182246 | 0.247464 | 0.270030 | -0.028737 | 0.162698 | 0.186835 | 0.027601 |
| total_deaths | 0.932079 | 1.000000 | 0.179812 | 0.394811 | 0.212619 | -0.031645 | 0.205128 | 0.198729 | 0.019990 |
| total_cases_pm | 0.182246 | 0.179812 | 1.000000 | 0.586468 | -0.056009 | 0.110043 | 0.313836 | 0.651200 | 0.081449 |
| total_deaths_pm | 0.247464 | 0.394811 | 0.586468 | 1.000000 | -0.013902 | 0.030281 | 0.389595 | 0.383672 | 0.120488 |
| population | 0.270030 | 0.212619 | -0.056009 | -0.013902 | 1.000000 | -0.023084 | 0.024395 | -0.059555 | -0.038329 |
| pop_density | -0.028737 | -0.031645 | 0.110043 | 0.030281 | -0.023084 | 1.000000 | 0.178878 | 0.315199 | 0.314973 |
| median_age | 0.162698 | 0.205128 | 0.313836 | 0.389595 | 0.024395 | 0.178878 | 1.000000 | 0.648905 | 0.662222 |
| gdp_per_capita | 0.186835 | 0.198729 | 0.651200 | 0.383672 | -0.059555 | 0.315199 | 0.648905 | 1.000000 | 0.296995 |
| hosp_beds | 0.027601 | 0.019990 | 0.081449 | 0.120488 | -0.038329 | 0.314973 | 0.662222 | 0.296995 | 1.000000 |
从上表可看出,总病例数与总死亡数之间的相关性非常高(0.93),而每百万人口总病例数与每百万人口总死亡数之间的相关性则较小(0.59),人均GDP与每百万人口总病例数之间也存在较强的相关关系(0.65)。
2.2 创建不同变量分位数的交叉表
首先创建一个DataFrame,使用qcut创建一个将数据分解为分位数的列,显示总病例数分位数与总死亡数数分位数的交叉表。
data_b['total_cases_q']=pd.qcut(data_b['total_cases'],labels=['very low','low','medium','high','very high'],q=5,precision=0)
data_b['total_deaths_q']=pd.qcut(data_b['total_deaths'],labels=['very low','low','medium','high','very high'],q=5,precision=0)
pd.crosstab(data_b.total_cases_q,data_b.total_deaths_q)| total_deaths_q | very low | low | medium | high | very high |
|---|---|---|---|---|---|
| total_cases_q | |||||
| very low | 34 | 7 | 1 | 0 | 0 |
| low | 12 | 19 | 10 | 1 | 0 |
| medium | 1 | 13 | 15 | 13 | 0 |
| high | 0 | 0 | 12 | 24 | 6 |
| very high | 0 | 0 | 2 | 4 | 36 |
data.loc[(data_b.total_cases_q=="very high")&(data_b.total_deaths_q=="medium")].T| iso_code | QAT | SGP |
|---|---|---|
| lastdate | 2020-06-01 | 2020-06-01 |
| location | Qatar | Singapore |
| total_cases | 56910 | 34884 |
| total_deaths | 38 | 23 |
| total_cases_pm | 19753.146 | 5962.727 |
| total_deaths_pm | 13.19 | 3.931 |
| population | 2881060.0 | 5850343.0 |
| pop_density | 227.322 | 7915.731 |
| median_age | 31.9 | 42.4 |
| gdp_per_capita | 116935.6 | 85535.383 |
| hosp_beds | 1.2 | 2.4 |
从表中可以看出,大多数国家都在对角线上或其附近,但有2个国家(卡塔尔,新加坡)的病例数很高,但死亡人数时中等的。
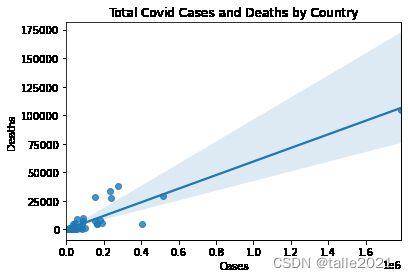
2.3 绘制两个变量间的散点图
使用seaborn的regplot方法时,除生成散点图外,还可以生成线性回归线。
import seaborn as sns
ax=sns.regplot(x="total_cases",y="total_deaths",data=data)
ax.set(xlabel='Cases',ylabel='Deaths',title='Total Covid Cases and Deaths by Country')
plt.show()# 检查回归线上方的意外值
data.loc[(data.total_cases<300000) & (data.total_deaths>20000)].T| iso_code | FRA | ITA | ESP | GBR |
|---|---|---|---|---|
| lastdate | 2020-06-01 | 2020-06-01 | 2020-05-31 | 2020-06-01 |
| location | France | Italy | Spain | United Kingdom |
| total_cases | 151753 | 233019 | 239429 | 274762 |
| total_deaths | 28802 | 33415 | 27127 | 38489 |
| total_cases_pm | 2324.879 | 3853.985 | 5120.952 | 4047.403 |
| total_deaths_pm | 441.251 | 552.663 | 580.197 | 566.965 |
| population | 65273512.0 | 60461828.0 | 46754783.0 | 67886004.0 |
| pop_density | 122.578 | 205.859 | 93.105 | 272.898 |
| median_age | 42.0 | 47.9 | 45.5 | 40.8 |
| gdp_per_capita | 38605.671 | 35220.084 | 34272.36 | 39753.244 |
| hosp_beds | 5.98 | 3.18 | 2.97 | 2.54 |
# 检查回归线下方的意外值
data.loc[(data.total_cases>400000) & (data.total_deaths<10000)].T| iso_code | RUS |
|---|---|
| lastdate | 2020-06-01 |
| location | Russia |
| total_cases | 405843 |
| total_deaths | 4693 |
| total_cases_pm | 2780.995 |
| total_deaths_pm | 32.158 |
| population | 145934460.0 |
| pop_density | 8.823 |
| median_age | 39.6 |
| gdp_per_capita | 24765.954 |
| hosp_beds | 8.05 |
从上表结果来看,有4个国家(法国,意大利,西班牙和英国)的病例数少于30w,死亡人数却超过2w;有1个国家(俄罗斯)的病例数超过30w,但死亡人数少于1w。
3. 使用线性回归来确定具有重大影响的数据点
使用线性回归的优势在于他们对于变量的分布依赖性较小,并且能比单变量或双变量分析揭示更多的东西,识别出在其它方面不显著的异常值。
使用statsmodels OLS方法拟合每百万人口总病例数的线性回归模型,然后确定那些对该模型具有较大影响的国家/地区。
3.1 描述性统计
xvars=['pop_density','median_age','gdp_per_capita']
data_x=data.loc[:,['total_cases_pm']+xvars].dropna()
data_x.describe()| total_cases_pm | pop_density | median_age | gdp_per_capita | |
|---|---|---|---|---|
| count | 175.000000 | 175.000000 | 175.000000 | 175.000000 |
| mean | 1134.015709 | 247.151863 | 30.537143 | 19008.385423 |
| std | 2101.363772 | 822.398967 | 9.117751 | 19673.386571 |
| min | 0.000000 | 1.980000 | 15.100000 | 661.240000 |
| 25% | 67.448000 | 36.066000 | 22.300000 | 4458.202500 |
| 50% | 263.413000 | 82.328000 | 29.700000 | 12951.839000 |
| 75% | 1357.506000 | 207.960000 | 38.700000 | 27467.146000 |
| max | 19753.146000 | 7915.731000 | 48.200000 | 116935.600000 |
3.2 拟合线性回归模型
def getlm(df):
Y=df.total_cases_pm
X=df[['pop_density','median_age','gdp_per_capita']]
X=sm.add_constant(X)
return sm.OLS(Y,X).fit()
lm=getlm(data_x)
lm.summary()| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 944.4731 | 426.712 | 2.213 | 0.028 | 102.172 | 1786.774 |
| pop_density | -0.2057 | 0.142 | -1.447 | 0.150 | -0.486 | 0.075 |
| median_age | -49.4398 | 16.013 | -3.088 | 0.002 | -81.048 | -17.832 |
| gdp_per_capita | 0.0921 | 0.008 | 12.015 | 0.000 | 0.077 | 0.107 |
3.3 确定对模型影响较大的国家/地区,并绘制影响图
在线性回归中,库克距离描述了单个样本对整个回归模型的影响程度,库克距离越大,说明影响越大。当库克距离大于0.5应仔细检查。
influence=lm.get_influence().summary_frame()
influence.loc[influence.cooks_d>0.5,['cooks_d']]| iso_code | cooks_d |
|---|---|
| HKG | 0.780662 |
| QAT | 5.080180 |
data_x.loc[influence.cooks_d>0.5]| total_cases_pm | pop_density | median_age | gdp_per_capita | |
|---|---|---|---|---|
| iso_code | ||||
| HKG | 0.000 | 7039.714 | 44.8 | 56054.92 |
| QAT | 19753.146 | 227.322 | 31.9 | 116935.60 |
fig,ax=plt.subplots(figsize=(12,8))
sm.graphics.influence_plot(lm,ax=ax,criterion="cooks")
plt.show()3.4 删除离群点,再进行回归拟合
data_x_new=data_x.loc[influence.cooks_d<0.5]
lm=getlm(data_x_new)
lm.summary()| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 44.0854 | 349.924 | 0.126 | 0.900 | -646.700 | 734.870 |
| pop_density | 0.2423 | 0.145 | 1.666 | 0.098 | -0.045 | 0.529 |
| median_age | -2.5165 | 13.526 | -0.186 | 0.853 | -29.217 | 24.184 |
| gdp_per_capita | 0.0557 | 0.007 | 7.875 | 0.000 | 0.042 | 0.070 |
删除之后,常数和median_age的估计值不再有效。回归输出的P>|t|值表明该系数是否与0有着显著不同。在第一次回归中,median_age和gdp_per_capita的系数在99%的置信水平上是显著的;即P>|t|小于0.01。而在删除两个离群值的情况下,只有gdp_per_capita是显著的。因此在处理此问题时,应分析诸如此类的离群值是否会增加重要信息或使模型失真。
4. 使用k最近邻算法找到离群值
在使用线性回归中,我们使用了每百万人口病例数作为因变量,而在没有标签数据的情况下,即没有目标变量或因变量时,无监督的机器学习工具也可以识别与其它观察结果不同的观察结果。
from pyod.models.knn import KNN
from sklearn.preprocessing import StandardScaler
# 创建分析列的标准化DataFrame
standardizer=StandardScaler()
var1=['location','total_cases_pm', 'total_deaths_pm', 'pop_density','median_age', 'gdp_per_capita']
data_var1=data.loc[:,var1].dropna()
data_var1_stand=standardizer.fit_transform(data_var1.iloc[:,1:])
# KNN模型生成异常分数
clf_name='KNN'
clf=KNN(contamination=0.1) #contamination:异常样本的百分比,在[0,0.5]之间的float型,默认为0.5
clf.fit(data_var1_stand)
y_pred=clf.labels_ # 返回训练数据上的分类标签 (0: 正常值, 1: 异常值)
y_scores=clf.decision_scores_ # 返回训练数据上的异常值 (分值越大越异常)
pred=pd.DataFrame(zip(y_pred,y_scores),columns=['outlier','scores'],index=data_var1.index)pred.outlier.value_counts()0 157 1 18 Name: outlier, dtype: int64
显示离群的数据
data_var1.join(pred).loc[pred.outlier==1,['location','total_cases_pm','total_deaths_pm','scores']].sort_values(['scores'],ascending=False)| location | total_cases_pm | total_deaths_pm | scores | |
|---|---|---|---|---|
| iso_code | ||||
| SGP | Singapore | 5962.727 | 3.931 | 9.228543 |
| HKG | Hong Kong | 0.000 | 0.000 | 7.770444 |
| QAT | Qatar | 19753.146 | 13.190 | 2.643709 |
| BHR | Bahrain | 6698.468 | 11.166 | 2.049642 |
| LUX | Luxembourg | 6418.776 | 175.726 | 1.525783 |
| MDV | Maldives | 3280.041 | 9.250 | 1.395496 |
| MLT | Malta | 1395.120 | 15.854 | 1.353541 |
| BGD | Bangladesh | 286.315 | 3.947 | 1.117904 |
| BRN | Brunei | 322.298 | 4.572 | 1.108011 |
| SAU | Saudi Arabia | 2449.053 | 14.448 | 0.902926 |
| ARE | United Arab Emirates | 3493.994 | 26.693 | 0.822334 |
| KWT | Kuwait | 6332.420 | 49.642 | 0.803449 |
| BRB | Barbados | 320.144 | 24.359 | 0.740434 |
| IRL | Ireland | 5060.962 | 334.562 | 0.721934 |
| PSE | Palestine | 122.907 | 0.980 | 0.717875 |
| NOR | Norway | 1551.489 | 43.532 | 0.712298 |
| GNQ | Equatorial Guinea | 930.872 | 8.553 | 0.695340 |
| OMN | Oman | 2239.641 | 9.204 | 0.670568 |
从上表可以看出,新加坡、卡塔尔的决策分数显著高于其它国家/地区,与前面的分析结果一致。
5. 使用隔离森林算法查找异常
隔离森林(isolation forest,也称孤立森林)找到离群点的方法是,对数据进行连续分区,直至某个数据点被隔离。如果某个数据点仅需要很少的分区即可隔离,那么它将获得较高的异常分数。
from sklearn.ensemble import IsolationForest
from mpl_toolkits.mplot3d import Axes3D
var2=['location','total_cases_pm', 'total_deaths_pm', 'pop_density','median_age', 'gdp_per_capita']
standardizer=StandardScaler()
data_var2=data.loc[:,var2].dropna()
data_var2_stand=standardizer.fit_transform(data_var2.iloc[:,1:])
clf=IsolationForest(n_estimators=100,max_samples='auto',contamination=0.1,max_features=1.0)
clf.fit(data_var2_stand)
data_var2['anomaly']=clf.predict(data_var2_stand) #查看隔离森林得出的结果,输出为一维矩阵,其中值为1代表为正常数据点,值为-1代表为异常点
data_var2['scores']=clf.decision_function(data_var2_stand) #数据集中样本的异常分数值,注意其中值是以0为阈值,小于0视为异常,大于0视为正常,data_var2.anomaly.value_counts()1 157 -1 18 Name: anomaly, dtype: int64
# 创建离群值和内围值的DataFrame
inlier,outlier=data_var2.loc[data_var2.anomaly==1],data_var2.loc[data_var2.anomaly==-1]
outlier[['location','total_cases_pm','total_deaths_pm','anomaly','scores']].sort_values(['scores']).head(10)| location | total_cases_pm | total_deaths_pm | anomaly | scores | |
|---|---|---|---|---|---|
| iso_code | |||||
| QAT | Qatar | 19753.146 | 13.190 | -1 | -0.257773 |
| SGP | Singapore | 5962.727 | 3.931 | -1 | -0.199578 |
| HKG | Hong Kong | 0.000 | 0.000 | -1 | -0.166863 |
| BEL | Belgium | 5037.354 | 816.852 | -1 | -0.116571 |
| BHR | Bahrain | 6698.468 | 11.166 | -1 | -0.110567 |
| LUX | Luxembourg | 6418.776 | 175.726 | -1 | -0.097430 |
| ITA | Italy | 3853.985 | 552.663 | -1 | -0.066504 |
| ESP | Spain | 5120.952 | 580.197 | -1 | -0.060739 |
| IRL | Ireland | 5060.962 | 334.562 | -1 | -0.040109 |
| MDV | Maldives | 3280.041 | 9.250 | -1 | -0.031414 |
ax=plt.axes(projection='3d')
ax.set_title('Isolation Forest Anomaly Detection')
ax.set_zlabel("Cases Per Million")
ax.set_xlabel("GDP Per Capita")
ax.set_ylabel("Median Age")
ax.scatter3D(inlier.gdp_per_capita,inlier.median_age,inlier.total_cases_pm,label="inlier",c='blue')
ax.scatter3D(outlier.gdp_per_capita,outlier.median_age,outlier.total_cases_pm,label="outlier",c='red')
ax.legend()
plt.tight_layout()
plt.show()隔离森林的结果与k最近邻的结果非常相似,卡塔尔、新加坡的异常得分较高(准确地说是最小负分),因此,当进行多变量分析时,可以考虑删除离群值再进行分析。