2021-07-07
YOLOV4训练自己的数据调优
数据集整理
- 网上的下载的数据集好多是XML格式的,而YOLO的数据集标签文件是txt格式的,这里提供方法将XML格式转为TXT格式的
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Sun Sep 15 21:56:32 2019
@author: youxinlin
"""
import copy
from lxml.etree import Element, SubElement, tostring, ElementTree
import xml.etree.ElementTree as ET
import pickle
import os
from os import listdir, getcwd
from os.path import join
import glob
# 当前目录
current_dir = os.path.dirname(os.path.abspath(__file__))
classes = ['单行车牌','双行车牌'] # 修改为自己类别
def convert(size, box):
dw = 1./size[0]
dh = 1./size[1]
x = (box[0] + box[1])/2.0
y = (box[2] + box[3])/2.0
w = box[1] - box[0]
h = box[3] - box[2]
x = x*dw
w = w*dw
y = y*dh
h = h*dh
return (x,y,w,h)
def convert_annotation(image_id):
in_file = open(os.path.join(current_dir, '%s.xml'%(image_id)))
out_file = open(os.path.join(current_dir, '%s.txt'%(image_id)),'w') #生成txt格式文件
tree=ET.parse(in_file)
root = tree.getroot()
size = root.find('size')
w = int(size.find('width').text)
h = int(size.find('height').text)
for obj in root.iter('object'):
cls = obj.find('name').text
if cls not in classes :
continue
for i in range(len(classes)):
if cls == classes[i] :
cls_id = i
xmlbox = obj.find('bndbox')
b = (float(xmlbox.find('xmin').text), float(xmlbox.find('xmax').text), float(xmlbox.find('ymin').text), float(xmlbox.find('ymax').text))
bb = convert((w,h), b)
out_file.write(str(cls_id) + " " + " ".join([str(a) for a in bb]) + '\n')
for pathAndFilename in glob.iglob("*.xml"):
title, ext = os.path.splitext(os.path.basename(pathAndFilename))
pathAndFilename=base_name=os.path.splitext(pathAndFilename)[0]
convert_annotation(pathAndFilename)
- 尽可能多的收集每一类的训练数据量,使得每种类型的样本分布均匀。
训练参数优化(修改cfg内容)
- batch越大越好,subdivisions越小越好,宽高越大越好,但相应的训练时间就越长,对资源要求就越高
[net]
batch=64 根据自己的gpu设置大小,理论上来说越大越好
subdivisions=64 将大batch分割为 subdivisions个小batch,每次读取 batch/subdivisions张图片计算loss,等到 subdivisions个小batch结束后更新总的loss,所以subdivisions尽可能小
# Training
width=608
height=608 图片的宽高越大越好,但必须为32的倍数,相应resize后的特征就会越多
#width=608
#height=608
- 训练次数 max_batches建议为classes*2000,不小于6000,但max_batches越大,效果越好,建议适当提高训练次数,直到loss趋于稳定,steps设为max_batches的80%-90%
learning_rate=0.0013
burn_in=20000
max_batches = 1000
policy=steps
steps=16000,18000
scales=.1,.1
我自己的训练对比图
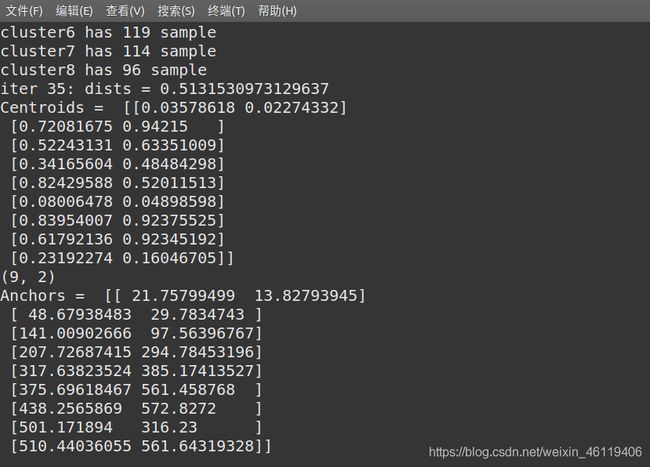
- 修改anchors的值,根据自己的数据集使用K-means++得到自己的预选框值
终端输入以下命令
python3 gen_anchors.py -filelist train.txt -num_clusters 9
将以下代码保存为文件gen_anchors.py
from os import listdir
from os.path import isfile, join
import argparse
#import cv2
import numpy as np
import sys
import os
import shutil
import random
import math
width_in_cfg_file = 608. //修改为自己设置的尺寸
height_in_cfg_file = 608.
def IOU(x,centroids):
similarities = []
k = len(centroids)
for centroid in centroids:
c_w,c_h = centroid
w,h = x
if c_w>=w and c_h>=h:
similarity = w*h/(c_w*c_h)
elif c_w>=w and c_h<=h:
similarity = w*c_h/(w*h + (c_w-w)*c_h)
elif c_w<=w and c_h>=h:
similarity = c_w*h/(w*h + c_w*(c_h-h))
else: #means both w,h are bigger than c_w and c_h respectively
similarity = (c_w*c_h)/(w*h)
similarities.append(similarity) # will become (k,) shape
return np.array(similarities)
def avg_IOU(X,centroids):
n,d = X.shape
sum = 0.
for i in range(X.shape[0]):
#note IOU() will return array which contains IoU for each centroid and X[i] // slightly ineffective, but I am too lazy
sum+= max(IOU(X[i],centroids))
return sum/n
def write_anchors_to_file(centroids,X,anchor_file):
f = open(anchor_file,'w')
anchors = centroids.copy()
print(anchors.shape)
for i in range(anchors.shape[0]):
anchors[i][0]*=width_in_cfg_file
anchors[i][1]*=height_in_cfg_file
widths = anchors[:,0]
sorted_indices = np.argsort(widths)
print('Anchors = ', anchors[sorted_indices])
for i in sorted_indices[:-1]:
f.write('%0.2f,%0.2f, '%(anchors[i,0],anchors[i,1]))
#there should not be comma after last anchor, that's why
f.write('%0.2f,%0.2f\n'%(anchors[sorted_indices[-1:],0],anchors[sorted_indices[-1:],1]))
f.write('%f\n'%(avg_IOU(X,centroids)))
print()
def kmeans(X,centroids,eps,anchor_file):
"""
X.shape = N * dim N代表全部样本数量,dim代表样本有dim个维度
centroids.shape = k * dim k代表聚类的cluster数,dim代表样本维度
"""
print("X.shape=",X.shape,"centroids.shape=",centroids.shape)
N = X.shape[0]
iterations = 0
k,dim = centroids.shape
prev_assignments = np.ones(N)*(-1)
iter = 0
old_D = np.zeros((N,k))
while True:
"""
D.shape = N * k N代表全部样本数量,k列分别为到k个质心的距离
1. 计算出D
2. 获取出当前样本应该归属哪个cluster
assignments = np.argmin(D,axis=1)
assignments.shape = N * 1 N代表N个样本,1列为当前归属哪个cluster
numpy里row=0,line=1,np.argmin(D,axis=1)即沿着列的方向,即每一行的最小值的下标
3. 将样本划分到相对应的cluster后,重新计算每个cluster的质心
centroid_sums.shape = k * dim k代表刻个cluster,dim列分别为该cluster的样本在该维度的均值
centroid_sums=np.zeros((k,dim),np.float)
for i in range(N):
centroid_sums[assignments[i]]+=X[i] # assignments[i]为cluster x 将每一个样本都归到其所属的cluster.
for j in range(k):
centroids[j] = centroid_sums[j]/(np.sum(assignments==j)) #np.sum(assignments==j)为cluster j中的样本总量
"""
D = []
iter+=1
for i in range(N):
d = 1 - IOU(X[i],centroids)
D.append(d)
D = np.array(D) # D.shape = (N,k)
print("iter {}: dists = {}".format(iter,np.sum(np.abs(old_D-D))))
assignments = np.argmin(D,axis=1)
#每个样本归属的cluster都不再变化了,就退出
if (assignments == prev_assignments).all() :
print("Centroids = ",centroids)
write_anchors_to_file(centroids,X,anchor_file)
return
#calculate new centroids
centroid_sums=np.zeros((k,dim),np.float)
for i in range(N):
centroid_sums[assignments[i]]+=X[i]
for j in range(k):
print("cluster{} has {} sample".format(j,np.sum(assignments==j)))
centroids[j] = centroid_sums[j]/(np.sum(assignments==j))
prev_assignments = assignments.copy()
old_D = D.copy()
def main(argv):
parser = argparse.ArgumentParser()
parser.add_argument('-filelist', default = '\\path\\to\\voc\\filelist\\train.txt',
help='path to filelist\n' )
parser.add_argument('-output_dir', default = 'generated_anchors/anchors', type = str,
help='Output anchor directory\n' )
parser.add_argument('-num_clusters', default = 0, type = int,
help='number of clusters\n' )
args = parser.parse_args()
if not os.path.exists(args.output_dir):
os.mkdir(args.output_dir)
f = open(args.filelist)
lines = [line.rstrip('\n') for line in f.readlines()]
#将label文件里的obj的w_ratio,h_ratio存储到annotation_dims
annotation_dims = []
for line in lines:
#line = line.replace('images','labels')
#line = line.replace('img1','labels')
line = line.replace('JPEGImages','labels')
line = line.replace('.jpg','.txt')
line = line.replace('.png','.txt')
print(line)
f2 = open(line)
for line in f2.readlines():
line = line.rstrip('\n')
w,h = line.split(' ')[3:]
#print(w,h)
annotation_dims.append(tuple(map(float,(w,h))))
annotation_dims = np.array(annotation_dims)
eps = 0.005
if args.num_clusters == 0:
for num_clusters in range(1,11): #we make 1 through 10 clusters
print(num_clusters)
anchor_file = join( args.output_dir,'anchors%d.txt'%(num_clusters))
indices = [ random.randrange(annotation_dims.shape[0]) for i in range(num_clusters)]
centroids = annotation_dims[indices]
kmeans(annotation_dims,centroids,eps,anchor_file)
print('centroids.shape', centroids.shape)
else:
anchor_file = join( args.output_dir,'anchors%d.txt'%(args.num_clusters))
##随机选取args.num_clusters个质心
indices = [ random.randrange(annotation_dims.shape[0]) for i in range(args.num_clusters)]
print("indices={}".format(indices))
centroids = annotation_dims[indices]
print("centroids=",centroids)
##
kmeans(annotation_dims,centroids,eps,anchor_file)
print('centroids.shape', centroids.shape)
if __name__=="__main__":
main(sys.argv)
[yolo]
mask = 0,1,2
#anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401
anchors = 30, 20, 92, 42, 145, 123, 216, 335, 223, 222, 325, 436, 416, 572, 490, 318, 496, 564
4.更改nms_kind,使用diounms
GreedyNMS是一种硬判决,是一种贪心算法,IOU是唯一考量的因素,选择一个好的阈值是多么重要的一件事,但又是一件很困难的事,经常会出现阈值设的小了,多个窗口融合为一个窗口,或者大了,一个物体检测出多种窗口。
基于此,DIOU-NMS就不仅仅考虑IOU,还考虑两个框中心点之间的距离。如果两个框之间IOU比较大,但是两个框的距离比较大时,可能会认为这是两个物体的框而不会被过滤掉。
iou_loss=ciou
nms_kind=diounms