igraph——图挖掘助力社会网络分析
http://www.ituring.com.cn/article/1762
社交网络(如Facebook,Twitter)可以完整地表现人们的生活。人们用不同的方式与他人互动,并且这些信息都可以在社交网络中抓取到。挖掘某个站点的有用信息可以帮助一些团体增加竞争力。
我最近无意中发现一款叫做“igraph”的工具,它提供了一些非常有效的挖掘功能。以下列举几条我觉得有意思的:
创建图表
图表由节点和连线组成,两者都可以附上一系列属性值(键/值对)。此外,连线可以是有向的也可以是无向的,还可以给它加上权重。
> library(igraph) > # Create a directed graph > g <- graph(c(0,1, 0,2, 1,3, 0,3), directed=T) > g Vertices: 4 Edges: 4 Directed: TRUE Edges: [0] 0 -> 1 [1] 0 -> 2 [2] 1 -> 3 [3] 0 -> 3 > # Create a directed graph using adjacency matrix > m <- matrix(runif(4*4), nrow=4) > m [,1] [,2] [,3] [,4] [1,] 0.4086389 0.2160924 0.1557989 0.2896239 [2,] 0.4669456 0.1071071 0.1290673 0.3715809 [3,] 0.2031678 0.3911691 0.5906273 0.7417764 [4,] 0.8808119 0.7687493 0.9734323 0.4487252 > g <- graph.adjacency(m > 0.5) > g Vertices: 4 Edges: 5 Directed: TRUE Edges: [0] 2 -> 2 [1] 2 -> 3 [2] 3 -> 0 [3] 3 -> 1 [4] 3 -> 2 > plot(g, layout=layout.fruchterman.reingold) >iGraph也提供了多种创建各种图形的图表的简单方法
> #Create a full graph > g1 <- graph.full(4) > g1 Vertices: 4 Edges: 6 Directed: FALSE Edges: [0] 0 -- 1 [1] 0 -- 2 [2] 0 -- 3 [3] 1 -- 2 [4] 1 -- 3 [5] 2 -- 3 > #Create a ring graph > g2 <- graph.ring(3) > g2 Vertices: 3 Edges: 3 Directed: FALSE Edges: [0] 0 -- 1 [1] 1 -- 2 [2] 0 -- 2 > #Combine 2 graphs > g <- g1 %du% g2 > g Vertices: 7 Edges: 9 Directed: FALSE Edges: [0] 0 -- 1 [1] 0 -- 2 [2] 0 -- 3 [3] 1 -- 2 [4] 1 -- 3 [5] 2 -- 3 [6] 4 -- 5 [7] 5 -- 6 [8] 4 -- 6 > graph.difference(g, graph(c(0,1,0,2), directed=F)) Vertices: 7 Edges: 7 Directed: FALSE Edges: [0] 0 -- 3 [1] 1 -- 3 [2] 1 -- 2 [3] 2 -- 3 [4] 4 -- 6 [5] 4 -- 5 [6] 5 -- 6 > # Create a lattice > g1 = graph.lattice(c(3,4,2)) > # Create a tree > g2 = graph.tree(12, children=2) > plot(g1, layout=layout.fruchterman.reingold) > plot(g2, layout=layout.reingold.tilford)iGraph还提供了另外两种图表生成的机制。“随机图表”可以在任意两个节点之间进行连线。而“优先连接”会给已经拥有较大度数的节点再增加连线(也就是多者更多)。
# Generate random graph, fixed probability > g <- erdos.renyi.game(20, 0.3) > plot(g, layout=layout.fruchterman.reingold, vertex.label=NA, vertex.size=5) # Generate random graph, fixed number of arcs > g <- erdos.renyi.game(20, 15, type='gnm') # Generate preferential attachment graph > g <- barabasi.game(60, power=1, zero.appeal=1.3)简单图表算法
这一节会介绍如何使用iGraph来实现一些简单的图表算法
最小生成树算法可以在图表里连接所有的节点,并使所有的连线权重最小。
# Create the graph and assign random edge weights > g <- erdos.renyi.game(12, 0.35) > E(g)$weight <- round(runif(length(E(g))),2) * 50 > plot(g, layout=layout.fruchterman.reingold, edge.label=E(g)$weight) # Compute the minimum spanning tree > mst <- minimum.spanning.tree(g) > plot(mst, layout=layout.reingold.tilford, edge.label=E(mst)$weight)连通分支算法可以找到会连通其他节点的连接,也就是说,两个节点之间的路径会穿过其他节点。需要注意的是,在无向图里连通是要对称的,在有向图(节点A指向节点B,但节点B不指向节点A的图表)里不是必须的。因此在有向图中存在一种连接的概念叫做“强”,也就是只有两个节点都分别指向对方才意味着它们是连通的。“弱”的连接意味着它们不是连通的。
> g <- graph(c(0, 1, 1, 2, 2, 0, 1, 3, 3, 4, 4, 5, 5, 3, 4, 6, 6, 7, 7, 8, 8, 6, 9, 10, 10, 11, 11, 9)) # Nodes reachable from node4 > subcomponent(g, 4, mode="out") [1] 4 5 6 3 7 8 # Nodes who can reach node4 > subcomponent(g, 4, mode="in") [1] 4 3 1 5 0 2 > clusters(g, mode="weak") $membership [1] 0 0 0 0 0 0 0 0 0 1 1 1 $csize [1] 9 3 $no [1] 2 > myc <- clusters(g, mode="strong") > myc $membership [1] 1 1 1 2 2 2 3 3 3 0 0 0 $csize [1] 3 3 3 3 $no [1] 4 > mycolor <- c('green', 'yellow', 'red', 'skyblue') > V(g)$color <- mycolor[myc$membership + 1] > plot(g, layout=layout.fruchterman.reingold)最短路径算法是最普遍的算法,它能找到节点A和节点B之间最短的路径。在iGraph里,如果图表是未加权的(也就是权重为1的)而且在权重为正时使用了迪杰斯特拉算法,会使用“breath-first search”算法。要是连线的权重是负数,则会使用Bellman-ford算法。
> g <- erdos.renyi.game(12, 0.25) > plot(g, layout=layout.fruchterman.reingold) > pa <- get.shortest.paths(g, 5, 9)[[1]] > pa [1] 5 0 4 9 > V(g)[pa]$color <- 'green' > E(g)$color <- 'grey' > E(g, path=pa)$color <- 'red' > E(g, path=pa)$width <- 3 > plot(g, layout=layout.fruchterman.reingold)图表统计
通过大量统计信息我们可以大致看到图表的形状。在最高权限下,我们可以看到图表的各类信息,它包括:
- 图表的大小(节点和连线的数量)
- 图表的密度是紧密的(|E|与|V|的平方成正比)还是稀疏的(|E|与|V|成正比)?
- 图表是连通的(大部分节点是互通的)还是非连通的(节点是孤立的)?
- 图表中最长的两点之间距离
- 有向图的对称性
- 出/入“度”的分布
> # Create a random graph > g <- erdos.renyi.game(200, 0.01) > plot(g, layout=layout.fruchterman.reingold, vertex.label=NA, vertex.size=3) > # No of nodes > length(V(g)) [1] 200 > # No of edges > length(E(g)) [1] 197 > # Density (No of edges / possible edges) > graph.density(g) [1] 0.009899497 > # Number of islands > clusters(g)$no [1] 34 > # Global cluster coefficient: > #(close triplets/all triplets) > transitivity(g, type="global") [1] 0.015 > # Edge connectivity, 0 since graph is disconnected > edge.connectivity(g) [1] 0 > # Same as graph adhesion > graph.adhesion(g) [1] 0 > # Diameter of the graph > diameter(g) [1] 18 > # Reciprocity of the graph > reciprocity(g) [1] 1 > # Diameter of the graph > diameter(g) [1] 18 > # Reciprocity of the graph > reciprocity(g) [1] 1 > degree.distribution(g) [1] 0.135 0.280 0.315 0.110 0.095 0.050 0.005 0.010 > plot(degree.distribution(g), xlab="node degree") > lines(degree.distribution(g))往下一点,我们也可以看到每对节点的统计信息,比如:
- 计算两点之间没有公用连线的路径(也就是需要移除多少条连线可以使两节点不连通)
- 计算两点之间的最短路径
- 计算两点之间路径的数量和长度
> # Create a random graph > g <- erdos.renyi.game(9, 0.5) > plot(g, layout=layout.fruchterman.reingold) > # Compute the shortest path matrix > shortest.paths(g) [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [1,] 0 1 3 1 2 2 1 3 2 [2,] 1 0 2 2 3 2 2 2 1 [3,] 3 2 0 2 1 2 2 2 1 [4,] 1 2 2 0 3 1 2 2 1 [5,] 2 3 1 3 0 3 1 3 2 [6,] 2 2 2 1 3 0 2 1 1 [7,] 1 2 2 2 1 2 0 2 1 [8,] 3 2 2 2 3 1 2 0 1 [9,] 2 1 1 1 2 1 1 1 0 > # Compute the connectivity matrix > M <- matrix(rep(0, 81), nrow=9) > M [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [1,] 0 0 0 0 0 0 0 0 0 [2,] 0 0 0 0 0 0 0 0 0 [3,] 0 0 0 0 0 0 0 0 0 [4,] 0 0 0 0 0 0 0 0 0 [5,] 0 0 0 0 0 0 0 0 0 [6,] 0 0 0 0 0 0 0 0 0 [7,] 0 0 0 0 0 0 0 0 0 [8,] 0 0 0 0 0 0 0 0 0 [9,] 0 0 0 0 0 0 0 0 0 > for (i in 0:8) { + for (j in 0:8) { + if (i == j) { + M[i+1, j+1] <- -1 + } else { + M[i+1, j+1] <- edge.connectivity(g, i, j) + } + } + } > M [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [1,] -1 2 2 3 2 3 3 2 3 [2,] 2 -1 2 2 2 2 2 2 2 [3,] 2 2 -1 2 2 2 2 2 2 [4,] 3 2 2 -1 2 3 3 2 3 [5,] 2 2 2 2 -1 2 2 2 2 [6,] 3 2 2 3 2 -1 3 2 3 [7,] 3 2 2 3 2 3 -1 2 3 [8,] 2 2 2 2 2 2 2 -1 2 [9,] 3 2 2 3 2 3 3 2 -1 > 中心性计算
在细节方面,我们可以看到各个节点的统计信息。根据这些数字可以测出节点的“中心性”
- 拥有较高出/入度数的节点也拥有较高的“度中心性”
- 与其他节点之间有短路径的节点拥有较高的“密集中心性”
- 与其他节点对之间有最短路径的节点拥有较高的“中间性”
- 连接了许多中心性较高节点的节点拥有较高的“特征向量中心性”
- 本地簇系数意味着相邻节点的互联性
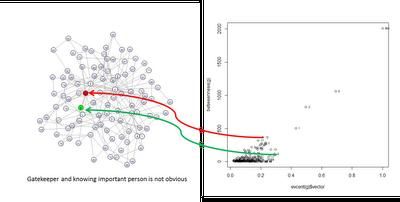
> # Degree > degree(g) [1] 2 2 2 2 2 3 3 2 6 > # Closeness (inverse of average dist) > closeness(g) [1] 0.4444444 0.5333333 0.5333333 0.5000000 [5] 0.4444444 0.5333333 0.6153846 0.5000000 [9] 0.8000000 > # Betweenness > betweenness(g) [1] 0.8333333 2.3333333 2.3333333 [4] 0.0000000 0.8333333 0.5000000 [7] 6.3333333 0.0000000 18.8333333 > # Local cluster coefficient > transitivity(g, type="local") [1] 0.0000000 0.0000000 0.0000000 1.0000000 [5] 0.0000000 0.6666667 0.0000000 1.0000000 [9] 0.1333333 > # Eigenvector centrality > evcent(g)$vector [1] 0.3019857 0.4197153 0.4197153 0.5381294 [5] 0.3019857 0.6693142 0.5170651 0.5381294 [9] 1.0000000 > # Now rank them > order(degree(g)) [1] 1 2 3 4 5 8 6 7 9 > order(closeness(g)) [1] 1 5 4 8 2 3 6 7 9 > order(betweenness(g)) [1] 4 8 6 1 5 2 3 7 9 > order(evcent(g)$vector) [1] 1 5 2 3 7 4 8 6 9从中Drew Conway发现拥有低“特征向量中心性”和高“中间性”的人是很重要联系人,而拥有高“特征向量中心性”和低“中间性”的人与重要的人有关联。现在我们来绘制“特征向量中心性”和“中间性”的图表。
> # Create a graph > g1 <- barabasi.game(100, directed=F) > g2 <- barabasi.game(100, directed=F) > g <- g1 %u% g2 > lay <- layout.fruchterman.reingold(g) > # Plot the eigevector and betweenness centrality > plot(evcent(g)$vector, betweenness(g)) > text(evcent(g)$vector, betweenness(g), 0:100, cex=0.6, pos=4) > V(g)[12]$color <- 'red' > V(g)[8]$color <- 'green' > plot(g, layout=lay, vertex.size=8, vertex.label.cex=0.6)在之后的帖子里我还会介绍一些特殊的社交网络分析的例子。
原文链接:Basic graph analytics using igraph(需要FQ)