python之实战----PCA、SVD、(NOnlinear PCA)KernelPCA、战iris
PCA
#-*- coding=utf-8 -*-
import numpy as np
from sklearn import datasets,decomposition,manifold
import matplotlib.pyplot as plt
def load_data():
iris=datasets.load_iris()
return iris.data,iris.target
def test_PCA(*data):
X,y=data
pca=decomposition.PCA(n_components=None)
pca.fit(X)
print('explained variance ratio : %s '%str(pca.explained_variance_ratio_))
if __name__=='__main__':
X,y=load_data()
test_PCA(X,y)
结果是四个特征值的比例4维度降到2维度:
PS E:\p> python test1.py
explained variance ratio : [ 0.92461621 0.05301557 0.01718514 0.00518309]pca画图
#-*- coding=utf-8 -*-
import numpy as np
from sklearn import datasets,decomposition,manifold
import matplotlib.pyplot as plt
def load_data():
iris=datasets.load_iris()
return iris.data,iris.target
def test_PCA(*data):
X,y=data
pca=decomposition.PCA(n_components=None)
pca.fit(X)
print('explained variance ratio : %s '%str(pca.explained_variance_ratio_))
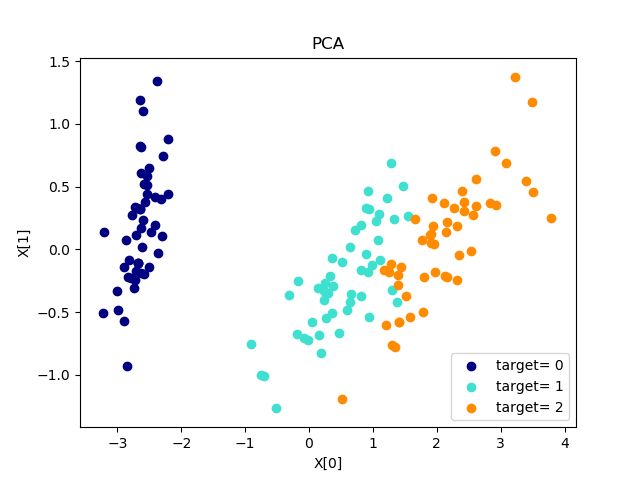
def plot_pca(*data):
X,y=data
pca=decomposition.PCA(n_components=2)#设置降到二维X[0],X[1]
pca.fit(X)
X_r=pca.transform(X)#将维度降低应用于X.
fig=plt.figure()
ax=fig.add_subplot(1,1,1)
'''
colors=((1,0,0),(0,1,0),(0,0,1),(0.5,0.5,0),(0,0.5,0.5),(0.5,0,0.5),
(0.4,0.6,0),(0.6,0.4,0),(0,0.6,0.4),(0.5,0.3,0.2),)
这里只用到前三个,因为我们知道鸢尾花数据集的特点
'''
colors = ['navy', 'turquoise', 'darkorange']
for label,color in zip(np.unique(y),colors):
#unique()保留数组中不同的值
position=y==label
ax.scatter(X_r[position,0],X_r[position,1],label="target= %d"%label,color=color)
ax.set_xlabel("X[0]")
ax.set_ylabel("X[1]")
ax.legend(loc="best")
ax.set_title("PCA")
plt.show()
if __name__=='__main__':
X,y=load_data()
plot_pca(X,y) 结果: