学习算法推荐
学习算法推荐系统
系统的总体构建思路是基于元学习的思想,基于给定的聚类任务特征(即元属性)进行聚类算法推荐,选择最优的聚类算法进行完成聚类分析。包括一个Gui图形用户界面,加载预处理.arff格式文件,对元学习算法进行参数设置,对推荐结果进行性能评估。元学习算法即一般的分类算法包括K-NN(最近邻算法)、ANN(人工神经网络)、C4.5(决策树)、SVM(支持向量机)、NB(朴素贝叶斯)、GA(基因算法)、Bagging(装袋融合算法)。元属性(即数据集的特征)包括基于简单度量的、汇总统计量的和信息论度量的。需要推荐的算法是聚类算法,相对于分类是一种无类标签指导的学习,在机器学习中也称为无监督学习,目前的元学习的研究主要用在分类任务上,对无监督学习聚类任务的研究国内外可供查阅的文献目前比较少,同时聚类任务在信息检索、web挖掘和蛋白质结构分析中有着特别重要的应用,如何从很多的聚类算法中选择一个适应任务特征的算法是当前的机器学习研究的一个重要方向。聚类算法主要包括:基于划分的、基于层次的、基于密度的、基于概率的、基于图模型的和基于融合的。具体系统构建思路:
1、做一个GUI,用于加载arff格式文件(是开源Java机器学习软件Weka中使用的格式),显示实验测试结果;
2、选择不同的聚类算法做为候选算法:KMeans、HierarchicalClusterer、DBSCAN、EM、ClusteringEnsemble、SCAN;
3、选择不同的元学习算法:C4.5、K-NN、NB、ANN、SVM、GA、Bagging;
4、选择不同的元属性:包括简单度量的、汇总统计量的和信息论度量的,在选择元属性时首先去除掉需要类标签信息的元属性;
5、选择实验数据集:数据集来自UCI机器学习中心机器学习中心数据库和开源Java机器学习软件Weka软件自带的arff属性关系格式文件;
6、选择聚类算法度量标准:包括无监督的内部度量和监督的外部度量;
7、选择元算法度量标准:使用分类算法的性能度量指标;
8、保存元算法的训练实例到MySQL数据库中,同时在推荐一次后将此次推荐的结果作为元实例存入数据库,增量的更新MySQL数据库;
构建的原型系统
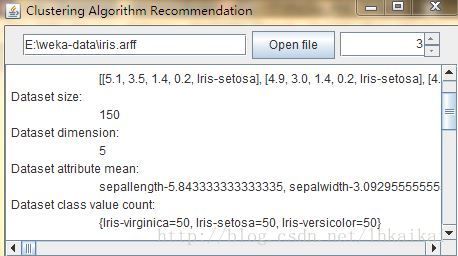
使用KMeans聚类算法进行系统平台测试,包括六个.java源文件,分别是Gui.java,ArffReader.java,KMeans.java,ClusteringEvaluation,java,UtilEnum.java(为Enum枚举类型),ArffFileFilter.java;测试数据集是iris.arff(Weka中自带的数据集)。源文件:java源文件压缩包;数据集:iris.arff源文件;可执行jar文件:jar可执行文件;如果主机没有安装Java运行环境至少需要安装一个JRE才可以运行可执行文件,官方网站中文:运行环境JRE;开源java机器学习软件Weka及其自带的.arff格式的数据集:Weka6.9,也可到官网http://www.cs.waikato.ac.nz/ml/weka/downloading.html下载。
1.Gui.java源代码
加载arff格式数据集,设置聚类算法参数,显示实验结果。
package clusteringAlgorithms;
import java.awt.BorderLayout;
import java.awt.Dimension;
import java.awt.Insets;
import java.awt.event.ActionEvent;
import java.awt.event.ActionListener;
import java.io.File;
import javax.swing.JButton;
import javax.swing.JFileChooser;
import javax.swing.JFrame;
import javax.swing.JPanel;
import javax.swing.JScrollPane;
import javax.swing.JSpinner;
import javax.swing.JTextArea;
import javax.swing.JTextField;
import javax.swing.SpinnerModel;
import javax.swing.SpinnerNumberModel;
import javax.swing.SwingUtilities;
import javax.swing.UIManager;
public class Gui extends JPanel implements ActionListener {
private static final long serialVersionUID = 001L;
JFileChooser jfc = new JFileChooser();
JButton jbt;
JTextArea log;
ArffReader arffReader;
JTextField url;
private static JSpinner spinner;
public Gui() {
super(new BorderLayout());
log = new JTextArea(10, 40);
log.setMargin(new Insets(5, 5, 5, 5));
log.setEditable(false);
JScrollPane logScrollPane = new JScrollPane(log);
jbt = new JButton("Open file");
jbt.addActionListener(this);
url = new JTextField(20);
SpinnerModel model = new SpinnerNumberModel(3, 1, Integer.MAX_VALUE , 1);
spinner = new JSpinner(model);
spinner.setPreferredSize(new Dimension(100, 25));
JPanel buttonPanel = new JPanel();
buttonPanel.add(url);
buttonPanel.add(jbt);
buttonPanel.add(spinner);
add(buttonPanel, BorderLayout.PAGE_START);
add(logScrollPane, BorderLayout.CENTER);
}
public void actionPerformed(ActionEvent e) {
if (e.getSource() == jbt) {
jfc.setDialogTitle("Open Arff File");
jfc.addChoosableFileFilter(new ArffFileFilter());
int returnval = jfc.showOpenDialog(Gui.this);
if (returnval == JFileChooser.APPROVE_OPTION) {
File file = jfc.getSelectedFile();
url.setText(file.getAbsolutePath());
arffReader = new ArffReader(file.getAbsolutePath());
log.append(arffReader.toString());
KMeans kMeans = new KMeans(Integer.parseInt(spinner.getValue().toString()));
log.append(kMeans.toString());
ClusteringEvaluation evaluation = new ClusteringEvaluation();
log.append(evaluation.toString());
}
log.setCaretPosition(log.getDocument().getLength());
}
}
private static void createAndShowGUI() {
JFrame frame = new JFrame("Clustering Algorithm Recommendation");
frame.setDefaultCloseOperation(JFrame.EXIT_ON_CLOSE);
frame.add(new Gui());
frame.pack();
frame.setVisible(true);
}
public static void main(String[] args) {
SwingUtilities.invokeLater(new Runnable() {
public void run() {
UIManager.put("swing.boldMetal", Boolean.FALSE);
createAndShowGUI();
}
});
}
}
2.ArffFileFilter.java源代码
过滤文件,只显示arff格式文件。
package clusteringAlgorithms;
import java.io.File;
public class ArffFileFilter extends javax.swing.filechooser.FileFilter {
@Override
public boolean accept(File f) {
// TODO Auto-generated method stub
return f.getAbsolutePath().endsWith(".arff") || f.isDirectory();
}
@Override
public String getDescription() {
// TODO Auto-generated method stub
return "*.arff";
}
}
3.ArffReader.java源代码
加载并预处理arff格式文件
package clusteringAlgorithms;
import java.io.BufferedReader;
import java.io.File;
import java.io.FileReader;
import java.io.IOException;
import java.io.PrintWriter;
import java.io.StreamTokenizer;
import java.nio.charset.Charset;
import java.nio.charset.StandardCharsets;
import java.nio.file.Path;
import java.nio.file.Paths;
import java.util.ArrayList;
import java.util.HashMap;
import java.util.Scanner;
import java.util.regex.Matcher;
import java.util.regex.Pattern;
import static clusteringAlgorithms.UtilEnum.*;
public class ArffReader {
private static String fileName = "E:\\weka-data\\iris.arff";
StreamTokenizer stoke;
ArrayList4.KMeans.java源代码
基于划分的聚类分析算法KMeans
package clusteringAlgorithms;
import java.util.ArrayList;
import java.util.HashMap;
import java.util.Iterator;
import java.util.Map;
import java.util.Random;
import static clusteringAlgorithms.UtilEnum.*;
public class KMeans {
private int cluNum = 2;
protected static ArrayList> dataset;
private ArrayList> nLabelDataset;
private ArrayList> centroids;
private HashMap> initC;
private int attNum;
private int numInsts;
private Random random;
private HashMap>> clusters;
private int numIterations;
protected static int[] clusterID;
public KMeans(int cluNum) {
this.cluNum = cluNum;
this.dataset = ArffReader.dataset;
numInsts = dataset.size();
removeClassLabel();
initializeCentroids();
buildClusters();
}
public ArrayList>
removeClassLabel() {
attNum = dataset.get(0).size();
nLabelDataset = new ArrayList>();
for (int i = 0; i < numInsts; i++)
nLabelDataset.add(new ArrayList());
for (int i = 0; i < dataset.size(); i++)
for (int j = 0; j < attNum - 1; j++)
nLabelDataset.get(i).add(Double.valueOf((String)dataset.get(i).get(j)));
attNum--;
return nLabelDataset;
}
public void initializeCentroids() {
int tempCentroid;
centroids = new ArrayList>();
random = new Random();
initC = new HashMap>();
for (int i = 0; i < cluNum; i++) {
tempCentroid = random.nextInt(numInsts);
while (initC.containsKey(tempCentroid))
tempCentroid = random.nextInt(numInsts);
initC.put(tempCentroid, nLabelDataset.get(tempCentroid));
centroids.add((ArrayList)nLabelDataset.get(tempCentroid).clone());
}
return;
}
public void buildClusters() {
double minDistance, temp;
int index;
ArrayList>[] cluster;
boolean isStillMoving;
clusterID = new int[numInsts];
isStillMoving = true;
index = 0;
cluster = new ArrayList[cluNum];
numIterations = 0;
//clusterID = new int[]
while (isStillMoving) {
clusters = new HashMap>>();
for (int i = 0; i < cluNum; i++)
cluster[i] = new ArrayList>();
isStillMoving = false;
for (int i = 0; i < numInsts; i++) {
minDistance = Double.POSITIVE_INFINITY;
for (int j = 0; j < cluNum; j++) {
temp = euclidDistance(nLabelDataset.get(i), centroids.get(j));
if (minDistance > temp) {
minDistance = temp;
index = j;
}
}
cluster[index].add(nLabelDataset.get(i));
clusters.put(Integer.valueOf(index), cluster[index]);
if (clusterID[i] != index) {
isStillMoving = true;
clusterID[i] = index;
}
}
moveCentroids();
numIterations++;
}
return;
}
public void moveCentroids() {
Integer key;
ArrayList> value;
double sum;
Iterator keyIterator;
keyIterator = clusters.keySet().iterator();
while (keyIterator.hasNext()) {
key = keyIterator.next();
value = clusters.get(key);
centroids.get(key).clear();
for (int i = 0; i < attNum; i++) {
sum = 0.0;
for (int j = 0; j < value.size(); j++)
sum += value.get(j).get(i);
sum /= value.size();
centroids.get(key).add(Double.valueOf(sum));
}
}
return;
}
public double euclidDistance(ArrayList point, ArrayList center) {
double distance, pointAtt, centerAtt;
distance = 0.0;
for (int i = 0; i < attNum; i++) {
pointAtt = point.get(i);
centerAtt = center.get(i);
distance += (pointAtt - centerAtt) * (pointAtt - centerAtt);
}
return distance;
}
public static int[] extractLabel(ArrayList> dataset) {
int[] labelArray;
int attNum, label;
label = 0;
attNum = dataset.get(0).size();
labelArray = new int[dataset.size()];
for (int i = 1; i < dataset.size(); i++) {
if (!dataset.get(i).get(attNum - 1).equals(dataset.get(i - 1).get(attNum - 1))) {
label++;
labelArray[i] = label;
}
else
labelArray[i] = label;
}
return labelArray;
}
public String toString() {
StringBuilder sb;
sb = new StringBuilder();
sb.append(NEWLINE.getConstant() +
"**********************KMeans clustering**********************" + NEWLINE.getConstant());
sb.append("Cluster number: " + NEWLINE.getConstant() + TAB.getConstant() +
cluNum + NEWLINE.getConstant());
sb.append("Initial centroids: " + NEWLINE.getConstant() + TAB.getConstant() +
initC + NEWLINE.getConstant());
sb.append("Total number of iterations: " + NEWLINE.getConstant() + TAB.getConstant() +
numIterations + NEWLINE.getConstant());
sb.append("Final centroids: " + NEWLINE.getConstant() + TAB.getConstant() +
centroids + NEWLINE.getConstant());
sb.append("Clustering result: " + NEWLINE.getConstant());
for (Map.Entry>> entry: clusters.entrySet()) {
sb.append(TAB.getConstant() + "Cluster " + entry.getKey() + ", size " +
entry.getValue().size() + ":" + NEWLINE.getConstant());
sb.append(TAB.getConstant() + TAB.getConstant() + "Cluster member: " +
entry.getValue() + NEWLINE.getConstant());
}
return sb.toString();
}
public static void main(String[] args) {
new ArffReader();
KMeans kMeans = new KMeans(3);
System.out.println(kMeans.toString());
}
}
5.ClusteringEvaluation.java源代码
使用了兰德指数和基于信息论的规范化的互信息性能度量标准,对聚类分析进行外部度量。
package clusteringAlgorithms;
import java.util.ArrayList;
import java.util.Arrays;
import java.util.Map;
import static clusteringAlgorithms.UtilEnum.*;
public class ClusteringEvaluation {
protected ArrayList> dataset;
protected int[] clusterID;
public ClusteringEvaluation() {
this.dataset = KMeans.dataset;
this.clusterID = KMeans.clusterID;
}
public static double randIndex(int[] cluRes, int[] truRes) {
double RI;
int ss, dd, sd, ds;
ss = dd = sd = ds = 0;
RI = 0.0;
if (cluRes.length != truRes.length)
return 0.0;
for (int i = 0; i < cluRes.length - 1; i++) {
for (int j = 0; j < cluRes.length; j++) {
if (cluRes[i] == cluRes[j] && truRes[i] == truRes[j])
ss++;
if (cluRes[i] != cluRes[j] && truRes[i] == truRes[j])
dd++;
if (cluRes[i] == cluRes[j] && truRes[i] != truRes[j])
sd++;
if (cluRes[i] != cluRes[j] && truRes[i] == truRes[j])
ds++;
}
}
RI = (double)(ss + dd) / (ss + dd + sd + ds);
return RI;
}
public static double log2(double value) {
return Math.log(value) / Math.log(2);
}
public static int uniqueEles(int[] array) {
int unique, size;
if (array == null)
return 0;
unique = 1;
size = array.length;
Arrays.sort(array);
for (int i = 0; i < size - 1; i++) {
if (array[i] == array[i+1])
continue;
else
unique++;
}
return unique;
}
public static int numOfElement(int ele, int[] array) {
int num;
num = 0;
for (int i: array)
if (ele == i)
num++;
return num;
}
public static int numOfIntersect(int x, int y, int[] a, int[] b) {
int num, size;
num = 0;
if (a.length != b.length)
return 0;
size = a.length;
for (int i = 0; i < size; i++)
if (a[i] == x && b[i] == y)
num++;
return num;
}
public static double mutuInfor(int[] cluRes, int[] truRes) {
int size, x, y;
double mi, px, py, pxy, hx, hy;
if (cluRes.length != truRes.length)
return 0.0;
size = cluRes.length;
mi = 0.0;
hx = 0.0;
hy = 0.0;
x = uniqueEles(cluRes);
y = uniqueEles(truRes);
for (int i = 0; i < x; i++) {
for (int j = 0; j < y; j++) {
px = (double)numOfElement(i, cluRes) / size;
py = (double)numOfElement(j, truRes) / size;
pxy = (double)numOfIntersect(i, j, cluRes, truRes) / size;
if (pxy != 0)
mi += pxy * log2(pxy / (px * py));
if (px != 0)
hx -= px * log2(px);
if (py != 0)
hy -= py * log2(py);
}
}
return (2 * mi) / (hx + hy);
}
public String toString() {
StringBuilder sb;
sb = new StringBuilder();
sb.append(NEWLINE.getConstant() +
"**********************Clustering Evaluation**********************" +
NEWLINE.getConstant());
sb.append("Rand index: " + NEWLINE.getConstant() + TAB.getConstant() +
randIndex(KMeans.clusterID, KMeans.extractLabel(KMeans.dataset)) +
NEWLINE.getConstant());
sb.append("Normal mutual information: " + NEWLINE.getConstant() + TAB.getConstant() +
mutuInfor(KMeans.clusterID, KMeans.extractLabel(KMeans.dataset)) +
NEWLINE.getConstant());
return sb.toString();
}
public static void main(String[] args) {
new ArffReader();
KMeans kMeans = new KMeans(3);
ClusteringEvaluation evaluation = new ClusteringEvaluation();
System.out.println(evaluation.toString());
}
}
6.UtilEnum.java
枚举类型文件,对在clusteringAlgorithms包中共同使用的字符串常量换行符“\n"和水平制表符”\t“进行枚举生成一个枚举文件。
package clusteringAlgorithms;
public enum UtilEnum {
NEWLINE("\n"), TAB("\t");
private String constant;
private UtilEnum(String s) {
constant = s;
}
public String getConstant() {
return constant;
}
}