医学图像预处理(五) 器官与病灶的直方图
事情起因是:
用模型训练分割肝脏,效果还不错。但是训练分割肝脏肿瘤时,dice系数很低。由于已经经过ROI处理,和图像预处理过程,所以只可能是数据层面出现了问题。经过查看,发现很多ct图是即使用肉眼也无法分辨出肿瘤的。论文中给出的那种图片,肿瘤与肝脏对比度很高,但这种情况只是数据集中的少数。为了验证自己的想法,在LITS2017的数据集上,做出了130个病人对应的肝脏与肿瘤的hu直方图,发现果然如此。
但这样的话,论文里是如何获得那么好的结果的呢??
(更新:2019-4-2,已完成肿瘤分割实验,写在博客:医学图像分割 基于深度学习的肝脏肿瘤分割 实战(二) )
下面是代码,可以作为工具类使用:
import numpy as np
import SimpleITK as sitk
import matplotlib.pyplot as plt
onServer = False
if onServer:
niiSegPath = './LITS17/seg/'
niiImagePath = './LITS17/ct/'
else:
niiSegPath = '~/LITS17/seg/'
niiImagePath = '~/LITS17/ct/'
def getRangeImageDepth(image):
z = np.any(image, axis=(1,2)) # z.shape:(depth,)
#print("all index:",np.where(z)[0])
if len(np.where(z)[0]) >0:
startposition,endposition = np.where(z)[0][[0,-1]]
else:
startposition = endposition = 0
return startposition, endposition
"""
会画出每个病人肿瘤区域最大切片的直方图
与汇总的直方图
"""
total_liver = []
total_tumor = []
colors = ['b','g']
for i in range(0, 131, 1):
seg = sitk.ReadImage(niiSegPath+ "segmentation-" + str(i) + ".nii", sitk.sitkUInt8)
segimg = sitk.GetArrayFromImage(seg)
src = sitk.ReadImage(niiImagePath+"volume-" + str(i) + ".nii")
srcimg = sitk.GetArrayFromImage(src)
seg_liver = segimg.copy()
seg_liver[seg_liver>0] = 1
seg_tumorimage = segimg.copy()
seg_tumorimage[segimg == 1] = 0
seg_tumorimage[segimg == 2] = 1
# 获取含有肿瘤切片的起、止位置
start,end = getRangeImageDepth(seg_tumorimage)
if start==0 and end == 0:
print("continue")
continue
print("start:",start," end:",end)
max_tumor=0 # 记录肿瘤的最大占比
max_tumor_index = -1 # 记录最大肿瘤所在切片
for j in range(start, end+1):
if np.mean(seg_tumorimage[j]) > max_tumor:
max_tumor = np.mean(seg_tumorimage[j])
max_tumor_index = j
# batch_image.append(srcimg[j,:,:])
# batch_mask.append(seg_tumorimage[j,:,:])
src_flatten = srcimg[max_tumor_index].flatten()
liver_flatten = seg_liver[max_tumor_index].flatten()
tumor_flatten = seg_tumorimage[max_tumor_index].flatten()
liver = [] # liver hu value
tumor = []
for j in range(src_flatten.shape[0]):
if liver_flatten[j]>0:
liver.append(src_flatten[j])
if tumor_flatten[j]>0:
tumor.append(src_flatten[j])
total_liver.append(liver)
total_tumor.append(tumor)
"""
# 因为肝脏区域很多而肿瘤很少,所以画直方图使用相同的y-axis就会导致
# 几乎看不到肿瘤的直方图
plt.hist(flat_total_liver, color = "skyblue", bins=100, alpha=0.5, label='liver hu')
plt.hist(flat_total_tumor, color = "red", bins=100, alpha=0.5, label='tumor hu')
plt.legend(loc='upper right')
"""
fig, ax1 = plt.subplots()
ax2 = ax1.twinx()
ax1.hist([liver, tumor], bins=100,color=colors)
n, bins, patches = ax1.hist([liver,tumor], bins=100)
ax1.cla() #clear the axis
#plots the histogram data
width = (bins[1] - bins[0]) * 0.4
bins_shifted = bins + width
ax1.bar(bins[:-1], n[0], width, align='edge', color=colors[0])
ax2.bar(bins_shifted[:-1], n[1], width, align='edge', color=colors[1])
#finishes the plot
ax1.set_ylabel("liver hu Count", color=colors[0])
ax2.set_ylabel("tumor hu Count", color=colors[1])
ax1.tick_params('y', colors=colors[0])
ax2.tick_params('y', colors=colors[1])
plt.tight_layout()
plt.title("person[%d],slice[%d]"%(i,max_tumor_index))
plt.savefig("LITS/person%d_slice%d.png"%(i,max_tumor_index))
plt.clf()
# 用来展平total_liver和total_tumor里面的值
flat_total_liver = [item for sublist in total_liver for item in sublist]
flat_total_tumor = [item for sublist in total_tumor for item in sublist]
#plt.hist(flat_total_liver, color = "skyblue", bins=100, alpha=0.5, label='liver hu')
#plt.hist(flat_total_tumor, color = "red", bins=100, alpha=0.5, label='tumor hu')
#plt.legend(loc='upper right')
#plt.title("total hu")
#plt.savefig("LITS/total.png")
#plt.clf()
fig, ax1 = plt.subplots()
ax2 = ax1.twinx()
ax1.hist([flat_total_liver, flat_total_tumor], bins=100,color=colors)
n, bins, patches = ax1.hist([flat_total_liver,flat_total_tumor], bins=100)
ax1.cla() #clear the axis
#plots the histogram data
width = (bins[1] - bins[0]) * 0.4
bins_shifted = bins + width
ax1.bar(bins[:-1], n[0], width, align='edge', color=colors[0])
ax2.bar(bins_shifted[:-1], n[1], width, align='edge', color=colors[1])
#finishes the plot
ax1.set_ylabel("liver hu Count", color=colors[0])
ax2.set_ylabel("tumor hu Count", color=colors[1])
ax1.tick_params('y', colors=colors[0])
ax2.tick_params('y', colors=colors[1])
plt.tight_layout()
plt.title("total")
plt.savefig("LITS/total.png")
plt.clf()
论文里的图片通常是这样的:
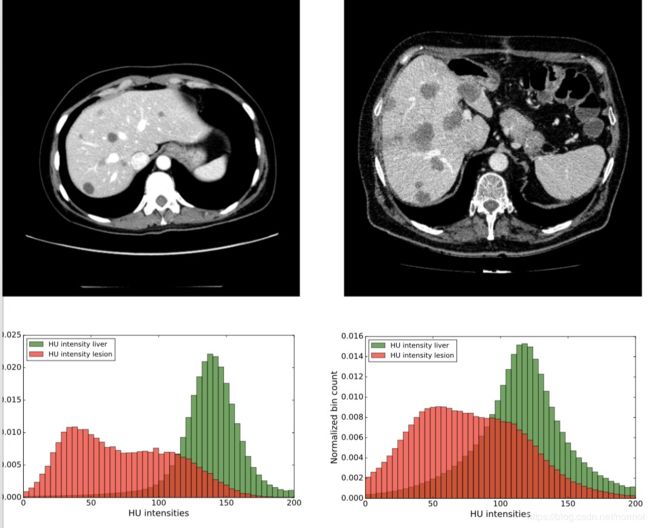
但实际上很多图是这样的(已经经过预处理):


为了验证想法,作出原始hu值,发现确实如此,大约一半的病人肝脏与肿瘤的hu分布是对比度很强的,但另一半的情况是二者几乎重叠。(问了下学医的同学,说肝脏肿瘤分为很多种,连医生很多时候也无法通过ct判断,这时候就要做增强ct)下面是代码运行的部分结果展示
