往NCBI上上传数据:
2.创建bioproject,以及biosample号
(Bioproject: Bioproject是一个项目的编号,在文章发表的时候,读者需要通过这个号来找到你文章中的数据存放到哪里去了,因此,在投稿之前,我们需要上传数据并拿到这个ID号。这个id号下面可以存放很多个不同的reads、序列等等,例如在我的项目里面,既有基因组的reads,基因组,也有重测序的reads,我可以把它全放到一个Bioproject下面,也可以申请两个bioproject号,一个用来放基因组的,一个用来放重测序的等等。
Biosample: Biosample与Bioproject类似,它是你所用的样本的一个ID。我们要先创建一个biosample,里面放有样本的信息。)
先创建Bioproject,再创建Biosample,按照页面的流程依次填写信息即可。
2.1 上传重测序的reads
2.1.1 创建Biosample
Attributes file 示例 (注,必填的信息一定要填,其他的随意)
*sample_name sample_title bioproject_accession *organism isolate cultivar ecotype age dev_stage *geo_loc_name *tissue biomaterial_provider cell_line cell_type collected_by collection_date culture_collection disease disease_stage genotype growth_protocol height_or_length isolation_source lat_lon phenotype population sample_type sex specimen_voucher temp treatment description
Pil01 Populus ilicifolia pil-01 not collected Kenya leaf
Pil02 Populus ilicifolia pil-02 not collected Kenya leaf
Pil03 Populus ilicifolia pil-03 not collected Kenya leaf
Pil04 Populus ilicifolia pil-04 not collected Kenya leaf
Pil05 Populus ilicifolia pil-05 not collected Kenya leaf
Pil06 Populus ilicifolia pil-06 not collected Kenya leaf
Pil07 Populus ilicifolia pil-07 not collected Kenya leaf
Pil08 Populus ilicifolia pil-08 not collected Kenya leaf
Pil09 Populus ilicifolia pil-09 not collected Kenya leaf
Pil10 Populus ilicifolia pil-10 not collected Kenya leaf
Pil11 Populus ilicifolia pil-11 not collected Kenya leaf
Pil12 Populus ilicifolia pil-12 not collected Kenya leaf
Pil13 Populus ilicifolia pil-13 not collected Kenya leaf
Pil14 Populus ilicifolia pil-14 not collected Kenya leaf
Pil15 Populus ilicifolia pil-15 not collected Kenya leaf
Pil16 Populus ilicifolia pil-16 not collected Kenya leaf
Pil17 Populus ilicifolia pil-17 not collected Kenya leaf
Pil18 Populus ilicifolia pil-18 not collected Kenya leaf
Pil19 Populus ilicifolia pil-19 not collected Kenya leaf 审核通过后,每个个体都会有一个Biosample ID号
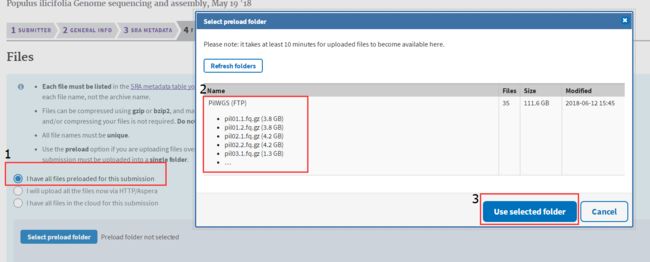
2.1.2 上传reads
选择(Sequence Read Archive)SRA上传reads
SRA meta file 示例:
bioproject_accession biosample_accession library_ID title library_strategy library_source library_selection library_layout platform instrument_model design_description filetype filename filename2 filename3 filename4 assembly
PRJNA471950 SAMN09228062 Pil01 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil01.1.fq.gz Pil01.2.fq.gz
PRJNA471950 SAMN09228063 Pil02 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil02.1.fq.gz Pil02.2.fq.gz
PRJNA471950 SAMN09228064 Pil03 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil03.1.fq.gz Pil03.2.fq.gz
PRJNA471950 SAMN09228065 Pil04 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil04.1.fq.gz Pil04.2.fq.gz
PRJNA471950 SAMN09228066 Pil05 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil05.1.fq.gz Pil05.2.fq.gz
PRJNA471950 SAMN09228067 Pil06 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil06.1.fq.gz Pil06.2.fq.gz
PRJNA471950 SAMN09228068 Pil07 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil07.1.fq.gz Pil07.2.fq.gz
PRJNA471950 SAMN09228069 Pil08 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil08.1.fq.gz Pil08.2.fq.gz
PRJNA471950 SAMN09228070 Pil09 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil09.1.fq.gz Pil09.2.fq.gz
PRJNA471950 SAMN09228071 Pil10 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil10.1.fq.gz Pil10.2.fq.gz
PRJNA471950 SAMN09228072 Pil11 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil11.1.fq.gz Pil11.2.fq.gz
PRJNA471950 SAMN09228073 Pil12 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil12.1.fq.gz Pil12.2.fq.gz
PRJNA471950 SAMN09228074 Pil13 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil13.1.fq.gz Pil13.2.fq.gz
PRJNA471950 SAMN09228075 Pil14 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil14.1.fq.gz Pil14.2.fq.gz
PRJNA471950 SAMN09228076 Pil15 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil15.1.fq.gz Pil15.2.fq.gz
PRJNA471950 SAMN09228077 Pil16 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil16.1.fq.gz Pil16.2.fq.gz
PRJNA471950 SAMN09228078 Pil17 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil17.1.fq.gz Pil17.2.fq.gz
PRJNA471950 SAMN09228079 Pil18 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil18.1.fq.gz Pil18.2.fq.gz
PRJNA471950 SAMN09228080 Pil19 WGS of P .ilicifolia WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil19.1.fq.gz Pil19.2.fq.gz 审核通过之后,就可以上传reads了!这里有两种方式:
- 用ftp从服务器直接上传:
如:
细叶杨的reads在:/share/work/01.all.reads/00.细叶杨populus_ilifolia这个目录下,操作如下:
cd /share/work/01.all.reads/00.细叶杨populus_ilifolia
lftp subftp:[email protected]
cd uploads/[email protected]_xxxx
mkdir PilWGS
cd PilWGS
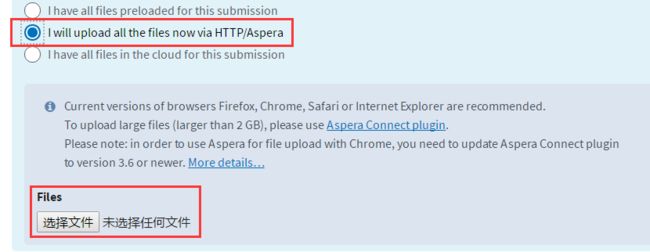
put pil02.1.fq.gz pil02.2.fq.gz ...- 用Aspera从电脑上上传:
下载并安装Asperea,要记住软件安装的目录
得到key文件并保存
在电脑打开cmd,进入aspera的bin目录
cd C:\Users\82002\AppData\Local\Programs\Aspera\Aspera Connect\bin
ascp -i G:\aspera.openssh -QT -l300m -k1 -d G:\PILWGS\ [email protected]:/uploads/[email protected]_xxxx/搞定了!最后就等着审核吧~
2.2 上传基因组序列
同样,创建Biosample号,然后选择Genome
其他的步骤省略,提交后,上传的基因组序列会进行审核,NCBI会发邮件告诉你通过还是有问题,需要继续修改。我就遇到了序列中含有线粒体序列以及接头的问题。。
放上修剪的脚本
#!/usr/bin/perl -w
use strict;
use Bio::SeqIO;
my $ip = "Pil.Genome.NCBI.fa";
my $op = "Pil.Genome.fa";
my $file = "02.Contamination.txt";
open (F,"<$file") or die ("$!\n");
open (O,">$op") or die ("$!\n");
my %exclude;
my %trim;
while (my $eve = ){
chomp ($eve);
#Exclude:
#Trim:
next if ($eve !~ /^scaffold/);
my @a = split /\s+/,$eve;
my $length = scalar (@a);
if ($eve !~ /\.\./){
my $chr = $a[0];
$exclude{$chr}++;
}elsif ($eve =~/\.\./){
my $chr = $a[0];
$exclude{$chr}++;
}elsif ($eve =~/\.\./){
my $chr = $a[0];
if ($a[2] !~ /,/){
$a[2] =~ /(\d+)\.\.(\d+)/;
my $start = $1;
my $end = $2;
$trim{$chr}{0}{start} = $start;
$trim{$chr}{0}{end} = $end;
}elsif ($a[2] =~ /,/){
my @b = split/,/,$a[2];
my $i=1;
foreach my $element (@b){
$element =~ /(\d+)\.\.(\d+)/;
my $start = $1;
my $end = $2;
$trim{$chr}{$i}{start} = $start;
$trim{$chr}{$i}{end} = $end;
$i++;
}
}
}
}
my $in=Bio::SeqIO->new(-file=>"$ip",-format=>'Fasta');
while(my $s =$in->next_seq()){
my $id=$s->id;
my $seq=$s->seq;
next if (exists $exclude{$id});
if (exists $trim{$id}){
my @a = split//,$seq;
foreach my $n (keys %{$trim{$id}}){
my $start = $trim{$id}{$n}{start};
my $end = $trim{$id}{$n}{end};
my $num = ($end-$start+1);
my $N = "N" x $num;
my @mv = split//,$N;
my @remove = splice (@a,$start-1,$end-$start+1,@mv);
}
my $newseq = join "", @a;
my $newseq_length = length ($newseq);
next if ($newseq_length < 200);
print O ">$id\n$newseq\n";
}
elsif ((! exists $exclude{$id}) && (! exists $trim{$id})){
print O ">$id\n$seq\n";
}
}
close F;
close O; 2.3 基因组reads的上传和重测序reads的上传类似,这里只放上填写的表格
Attributes file
accession sample_name sample_title bioproject_accession organism isolate cultivar ecotype age dev_stage geo_loc_name tissue biomaterial_provider cell_line cell_type collected_by collection_date culture_collection disease disease_stage genotype growth_protocol height_or_length isolation_source lat_lon phenotype population sample_type sex specimen_voucher temp treatment description
SAMN09388354 Pil500 Populus ilicifolia Pil-500 not collected kenya leaf
SAMN09388355 Pil800 Populus ilicifolia Pil-800 not collected kenya leaf
SAMN09388356 Pil2k Populus ilicifolia Pil-2k not collected kenya leaf
SAMN09388357 Pil5k Populus ilicifolia Pil-5k not collected kenya leaf
SAMN09388358 Pil10k Populus ilicifolia Pil-10k not collected kenya leaf SRA meta file
bioproject_accession sample_name library_ID title library_strategy library_source library_selection library_layout platform instrument_model design_description filetype assembly filename filename2 filename3 filename4 filename5 filename6 filename7 filename8
PRJNA471950 Pil500 Pil-500 WGS of P .ilicifolia for assemble WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil.genome.500bp.1.fq.gz Pil.genome.500bp.2.fq.gz
PRJNA471950 Pil800 Pil-800 WGS of P .ilicifolia for assemble WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil.genome.800bp.1.fq.gz Pil.genome.800bp.2.fq.gz
PRJNA471950 Pil2k Pil-2k WGS of P .ilicifolia for assemble WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil.genome.2kbp.1.fq.gz Pil.genome.2kbp.2.fq.gz
PRJNA471950 Pil5k Pil5k WGS of P .ilicifolia for assemble WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil.genome.5kbp.1.fq.gz Pil.genome.5kbp.2.fq.gz
PRJNA471950 Pil10k Pil-10k WGS of P .ilicifolia for assemble WGS GENOMIC RANDOM paired ILLUMINA HiSeq X Ten leaves used to extract genomic DNA and WGS Illumina protocol fastq Pil.genome.10kbp.1.fq.gz Pil.genome.10kbp.2.fq.gz ---END--