- python做生物信息学分析_Python从零开始第五章生物信息学①提取差异基因
吴敬欣
python做生物信息学分析
目前来说,做生物信息学的人越来越多,但是我觉得目前而言做生信的主要有三类人:老本行是做实验的,做生信可能是为了辅助研究或者是为了发paper(有非常多的临床生选择趟生信这波水)主要是做生信的,主要涵盖高通量测序数据分析,组学数据分析等等,专门从事生物学数据分析的这群人,其大部分也是本科生物狗作为强大的生力军,以调包写R,python为主。那么这群人就要熟悉看各种包的tutorial以及如何进行常规
- 用Python实现生信分析——功能预测详解
写代码的M教授
生信分析python开发语言
功能预测是生物信息学中的一项重要任务,通过分析基因或蛋白质序列的特征,推测它们的生物学功能。功能预测通常涉及多种方法,包括序列比对、基序识别、机器学习模型等。这些方法可以帮助科学家推断未知基因的功能,从而加速生物学研究的进展。1.功能预测的主要方法(1)同源性比对:通过将未知基因或蛋白质序列与数据库中的已知序列进行比对,识别出同源序列,并推测它们的功能。常用工具包括BLAST、HMMER等。(2)
- 用Python实现生信分析——序列搜索和比对工具详解
写代码的M教授
生信分析python
1.什么是序列搜索和比对工具?序列搜索和比对工具在生物信息学中用于在大型序列数据库中搜索与查询序列相似的序列,并进行比对分析。这些工具可以帮助研究人员识别与目标序列相关的已知序列,从而推测其功能、结构和进化关系。常见的序列搜索和比对工具包括:BLAST(BasicLocalAlignmentSearchTool):最常用的序列搜索工具,能够快速找到与查询序列相似的序列。FASTA:另一个常用的序列
- 大模型在生物信息学中的应用前景
AI天才研究院
AI人工智能与大数据ChatGPTjavapythonjavascriptkotlingolang架构人工智能大厂程序员硅基计算碳基计算认知计算生物计算深度学习神经网络大数据AIGCAGILLM系统架构设计软件哲学Agent程序员实现财富自由
大模型在生物信息学中的应用前景关键词:大模型、生物信息学、基因组学、蛋白质组学、应用前景摘要:本文将深入探讨大模型在生物信息学中的应用前景。首先,我们将介绍大模型的基础知识,包括其定义、特点和优势。接着,我们将分析大模型在生物信息学中的问题背景和具体应用场景。然后,我们将详细讲解大模型在生物信息学中的数据处理与分析方法,以及其在基因组学和蛋白质组学中的应用案例。最后,我们将讨论大模型在生物信息学中
- 【深度学习】条件随机场(CRF)深度解析:原理、应用与前沿
白熊188
深度学习深度学习人工智能
条件随机场(CRF)深度解析:原理、应用与前沿一、算法背景知识1.1序列标注的挑战1.2概率图模型演进二、算法理论与结构2.1基本定义2.2特征函数设计状态特征(节点特征)转移特征(边特征)2.3线性链CRF结构2.4训练与解码2.5前向-后向算法三、模型评估3.1评估指标3.2评估方法对比3.3性能基准(CoNLL-2003NER)四、应用案例4.1自然语言处理4.2生物信息学4.3计算机视觉五
- 最新期刊影响因子,基本包含全部期刊
Bioinfo科研生信筆記
影响因子2024年期刊影响因子期刊因子因子IF
原文链接:2024年期刊最新影响因子(IF)2024年期刊最新影响因子(IF)BioinfoR生信筆記,注于分享生物信息学相关知识和R语言绘图教程。
- 向量检索中的 ANN(Approximate Nearest Neighbor)技术
XiaoQiong.Zhang
AI人工智能
向量检索中的ANN(ApproximateNearestNeighbor)技术是一种在高维空间中高效查找与查询向量q最相似的Top-K个向量的方法,其核心在于牺牲一定的精度(召回率)以换取比精确最近邻搜索(ExactNN)高数个数量级的查询速度。它广泛应用于图像/视频检索、自然语言处理(如语义搜索、问答)、推荐系统、生物信息学等场景。⸻一、基本问题定义目标:给定一个查询向量q,在一个庞大的向量集合
- cd-hit安装与使用-cd-hit v4.8.1(bioinfomatics tools-005)
让学习成为一种生活方式
基因组多组学序列比对githublinux论文阅读数据挖掘
01背景介绍CD-HIT(ClusterDatabaseatHighIdentitywithTolerance)是一种广泛使用的生物信息学工具,主要用于快速聚类生物序列数据,如蛋白质或核酸序列,以减少数据冗余和简化数据分析。其基本原理涉及比较序列之间的相似性,将高度相似的序列分组到同一个聚类中,从而减少数据集的复杂性。1.1算法原理CD-HIT的算法原理主要包括以下几个方面:序列比较和相似性评分:
- 基于 Java 的大数据分布式计算在基因编辑数据分析与精准医疗中的应用进展
知识产权13937636601
计算机java分布式计算基因编辑
随着基因测序成本断崖式下降(单人类全基因组低于100)和CRISPR基因编辑技术成熟,全球日均产生超20PB基因数据。传统单机生物信息学工具难以应对海量多组学数据的整合、分析与临床转化。本文将系统阐述**Java技术栈如何构建新一代基因大数据计算中枢**:基于Hadoop+Spark的分布式架构实现千倍加速的基因组比对;通过Flink流式计算引擎支撑CRISPR脱靶效应实时预测;利用ApacheA
- PostgreSQL 在生物信息学中的应用
belldeep
PostgreSQL生物信息学postgresql数据库生物信息学
PostgreSQL(简称PG)是一种强大的开源关系型数据库管理系统,因其高可靠性、扩展性和支持复杂查询的特性,在生物信息学领域得到广泛应用。以下是其核心应用场景及优势分析:一、生物数据存储与管理生物信息学涉及海量异构数据,PG的结构化存储能力和可扩展性使其成为理想选择。1.多类型数据存储基因组数据:存储DNA/RNA序列、基因注释(如GTF/GFF文件)、变异数据(VCF格式)等。例:将基因组序
- 一款适合程序员的流程图/思维导图利器
qq_21478261
#Python可视化python运维思维导图图论机器学习
首发地址:程序员必备流程图/思维导图利器本文介绍graphviz在Python中的接口。graphviz是在复杂网络、生物信息学、软件工程、数据库和网页设计、机器学习等领域使用广泛的图(Graph)可视化利器。graphviz支持Linux、Windows、Mac、Solaris等多个系统,拥有多种编程语言的API(perl、python、ruby、C#等)。graphviz功能先看看graphv
- 支持向量机SVM:从数学原理到实际应用
代码很孬写
支持向量机算法机器学习语言模型自然语言处理ai人工智能
前言本篇文章全面深入地探讨了支持向量机(SVM)的各个方面,从基本概念、数学背景到Python和PyTorch的代码实现。文章还涵盖了SVM在文本分类、图像识别、生物信息学、金融预测等多个实际应用场景中的用法。一、引言背景支持向量机(SVM,SupportVectorMachines)是一种广泛应用于分类、回归、甚至是异常检测的监督学习算法。自从Vapnik和Chervonenkis在1995年首
- 7天掌握!MySQL vs 图数据库:混合架构下的复杂关系分析全揭秘
墨瑾轩
数据库学习数据库mysql架构
关注墨瑾轩,带你探索编程的奥秘!超萌技术攻略,轻松晋级编程高手技术宝库已备好,就等你来挖掘订阅墨瑾轩,智趣学习不孤单即刻启航,编程之旅更有趣在当今的数据密集型世界中,处理和理解复杂的关系网络变得越来越重要。从社交网络到推荐系统,从生物信息学到金融风险评估,这些领域都需要一种能够高效处理高度互联数据的技术。传统的关系型数据库如MySQL,在处理这类问题时遇到了瓶颈。而图数据库则以其独特的结构优势脱颖
- 汉明距离(Hamming Distance)
追逐此刻
算法方法python算法开发语言
1.定义汉明距离是指两个等长字符串在相同位置上不同字符的个数。它常用于衡量两个字符串的相似度,广泛应用于编码理论、信息论、密码学、生物信息学等领域。2.数学表达给定两个等长的字符串x和y,汉明距离d(x,y)定义为:其中:n是字符串的长度,xi和yi分别是x和y的第i个字符,Ⅱ(⋅)是指示函数(当条件成立时返回1,否则返回0)。3.示例二进制字符串:x="10110",y="11110"比较每一位
- Newcpgreport:CpG岛甲基化差异分析
简说基因-专业生信合作伙伴
在人类基因组中,约60%的基因启动子区域都蕴藏着特殊的DNA序列——CpG岛。CpG岛(富含CpG二核苷酸的区域)被称为基因调控的“开关”,它们常位于基因启动子区域,与DNA甲基化、基因沉默等表观遗传现象密切相关。要精准定位这些区域,生物信息学家们开发了多种工具,其中newcpgreport凭借其独特的算法设计和可靠的检测性能,成为该领域的明星工具。功能特点核心功能与原理1.滑动窗口检测法newc
- 生物医学工程导论:学习笔记(四)
Zodornus
生物医学工程学习笔记
生物信息学(Bioinformatics)狭义概念:应用信息科学的理论、方法和技术,来管理、分析和利用生物分子数据。广义概念:应用信息科学的方法和技术,研究生物体系和生物过程中信息的存储、信息的内涵和信息的传递,研究和分析生物体细胞、组织、器官的生理、病理、药理过程中的各种生物信息。(生命科学中的信息科学)目的:处理、归纳、总结海量的生物实验数据,并找到其中的规律。成果:基因测序等。研究内容基因组
- 探索生物信息学的未来:Rust-Bio 库
富嫱蔷
探索生物信息学的未来:Rust-Bio库rust-bioThislibraryprovidesimplementationsofmanyalgorithmsanddatastructuresthatareusefulforbioinformatics.Allprovidedimplementationsarerigorouslytestedviacontinuousintegration.项目地址
- 2025.04.18【数据修复】DCA:高效缺失值插补工具解析
穆易青
单细胞信息可视化
文章目录1.DCA工具简介2.DCA的安装方法3.DCA常用命令1.DCA工具简介在生物信息学领域,数据分析是一个复杂且耗时的过程。DCA(DifferentialCorrelationAnalysis)工具是一个专门设计来识别和分析差异相关性的统计工具。它能够帮助研究者从大量的生物医学数据中,发现变量间的相关性变化,这对于理解复杂疾病的分子机制至关重要。DCA工具通过计算和比较不同样本或条件下变
- 2025.04.17【Stacked area】| 生信数据可视化:堆叠区域图深度解析
穆易青
信息可视化
文章目录生信数据可视化:堆叠区域图深度解析堆叠面积图简介为什么使用堆叠面积图如何使用R语言创建堆叠面积图安装和加载ggplot2包创建堆叠面积图的基本步骤示例代码解读堆叠面积图堆叠面积图的局限性实际应用案例示例:基因表达量随时间变化结论生信数据可视化:堆叠区域图深度解析在生物信息学领域,数据可视化是理解复杂数据集的关键。其中,堆叠面积图(StackedAreaChart)是一种展示多个群体随时间变
- DNA、蛋白质、生物语义语言模型的介绍
bug开发工程师.
语言模型人工智能自然语言处理
主要模型概述ProtBERT:专注于蛋白质序列嵌入,支持多种下游任务如序列分类和功能预测。ProtGPT2:利用生成式模型生成高质量的蛋白质序列,适用于新蛋白质设计。AlphaFold:革命性地预测蛋白质三维结构,推动了结构生物学的发展。TAPE:提供统一的框架进行蛋白质序列表示学习,支持多种生物信息学任务。BioBERT:针对生物医学文本挖掘设计的模型,提升了生物信息处理能力。DNA-BERT:
- matlab在生物学中的应用,MATLAB在生物信息学分析中的应用.doc
weixin_39599097
matlab在生物学中的应用
MATLAB在生物信息学分析中的应用MATLAB在生物序列信息分析中的应用生物技术(生物制药方向09)杨清松0909501162摘要:MATLAB生物信息工具箱为广大用户提供了一个用于基因组和蛋白质组分析的综合环境,它利用数据库资源,使科学研究事半功倍,在工具箱提供的开放环境里,用户甚至可以按照自己的目的来设计和利用分析工具。本文主要介绍MATLAB生物信息工具箱在基因序列分析中的应用,包括确定核
- 用Python实现生信分析——隐马尔可夫模型(HMM)在生物信息学中的应用详解
写代码的M教授
生信分析人工智能python
在生物信息学中,隐马尔可夫模型(HMM)被广泛应用于基因组注释、蛋白质结构预测、基因预测等领域。以下是针对生物信息学应用的详细讲解,包括案例、Python实现、运行结果和分析。1.HMM在生物信息学中的应用场景HMM在生物信息学中的应用非常广泛,以下是一些典型场景:(1)基因预测:HMM可以用来预测DNA序列中的基因。通过建模不同区域(如外显子、内含子、启动子等)的特征,HMM可以识别出可能的基因
- 生物信息学数据库分类
划过手的泪滴t
生物信息学数据库
生物信息学数据库(一)文献数据库1、PubMed:拥有超过两百六十万生物医学文献的数据库,这些文献来源于MEDLINE,也就是生物医学文献数据库、生命科学领域学术杂志、以及在线的专业书籍。链接:PubMed(nih.gov)PubMed存在的问题(1)搜索1995年前文献中排名是为以后的作者(2)搜索1976年以前的文献是没有摘要的(3)1965年前的文献较难搜索(二)一级核酸数据库1、※GenB
- 生物信息学技能树(Bioinformatics)与学习路径
lisw05
生物信息学生物信息学
李升伟整理生物信息学是一门跨学科领域,涉及生物学、计算机科学以及统计学等多个方面。以下是关于生物信息学的学习路径及相关技能的详细介绍。一、基础理论知识1.生物学基础知识需要掌握分子生物学、遗传学、细胞生物学等相关概念。对基因组结构、蛋白质功能及其相互作用有基本理解。2.编程能力掌握至少一种脚本语言(如Python或Perl),用于数据处理和自动化任务3。学习R语言进行数据分析和可视化。3.统计学与
- centos-LLM-生物信息-BioGPT-使用1
淀粉肠狂热粉
生物信息学centoslinux生信生物信息AIGC
参考:GitHub-microsoft/BioGPThttps://github.com/microsoft/BioGPTBioGPT:用于生物医学文本生成和挖掘的生成式预训练转换器|生物信息学简报|牛津学术—BioGPT:generativepre-trainedtransformerforbiomedicaltextgenerationandmining|BriefingsinBioinfor
- 【机器学习】每日一讲-朴素贝叶斯公式
问道飞鱼
机器学习与人工智能机器学习人工智能朴素贝叶斯公式
文章目录**一、朴素贝叶斯公式详解****1.贝叶斯定理基础****2.从贝叶斯定理到分类任务****3.特征独立性假设****4.条件概率的估计****二、在AI领域的作用****1.文本分类与自然语言处理(NLP)****2.推荐系统****3.医疗与生物信息学****4.实时监控与异常检测****5.多模态数据处理****三、推导过程示例(以文本分类为例)****四、代码实现(Python)
- 2025.04.08【工具探索】| SC3:交互式聚类分析的新纪元
穆易青
ClusteringInteractive
文章目录1.SC3工具简介:探索生物信息学中的聚类分析利器1.1为什么选择SC3?1.2SC3的主要功能2.SC3的安装方法:轻松步入单细胞数据分析的大门2.1安装R语言环境2.2安装SC3包2.3安装依赖包3.SC3常用命令:掌握高效数据分析的钥匙3.1数据预处理3.2特征选择3.3聚类分析3.4结果可视化3.5高级分析4.SC3的案例研究4.1数据获取4.2数据预处理和特征选择4.3聚类分析4
- 2025.04.09【Sankey】| 生信数据流可视化精讲
穆易青
R语言信息可视化
文章目录引言Sankey图简介R语言中的Sankey图实现安装和加载`networkD3`包创建Sankey图的数据结构创建Sankey图绘制Sankey图结论引言在生物信息学领域,数据可视化是理解和分析复杂数据集的关键工具之一。今天,我们将深入探讨一种特别适用于展示数据流动的图表——Sankey图。这种图表通过矩形或文本表示实体(节点),并使用箭头或弧线显示实体间的流动,非常适合展示能量或成本的
- 生信数据集解读记录DrugBank,ATC分类系统
混入生信圈的yu
人工智能
最近在阅读Improvingdrug–druginteractionspredictionwithinterpretabilityviameta-path-basedinformationfusion看数据集,记录一些不懂的地方。关于DrugBank:DrugBank数据库是一个综合性的生物信息学和化学信息学资源,它提供了详细的药物数据和全面的药物目标信息。它包含的数据类型非常广泛,包括但不限于:
- 2025.04.08【技术分享】| bseqsc:一站式解决差异表达分析、数据整合与可视化挑战
穆易青
IntegrationVisualisation
文章目录1.bseqsc工具简介:探索基因表达的微观世界2.bseqsc的安装方法:搭建你的生物信息学工作台系统要求安装步骤常见问题解决策略3.bseqsc常用命令:掌握基因表达分析的钥匙数据预处理差异表达分析结果可视化高级分析结论1.bseqsc工具简介:探索基因表达的微观世界在生物信息学领域,基因表达分析是一个核心任务,它帮助我们理解基因如何在不同条件下发挥作用。bseqsc工具便是这样一个强
- rust的指针作为函数返回值是直接传递,还是先销毁后创建?
wudixiaotie
返回值
这是我自己想到的问题,结果去知呼提问,还没等别人回答, 我自己就想到方法实验了。。
fn main() {
let mut a = 34;
println!("a's addr:{:p}", &a);
let p = &mut a;
println!("p's addr:{:p}", &a
- java编程思想 -- 数据的初始化
百合不是茶
java数据的初始化
1.使用构造器确保数据初始化
/*
*在ReckInitDemo类中创建Reck的对象
*/
public class ReckInitDemo {
public static void main(String[] args) {
//创建Reck对象
new Reck();
}
}
- [航天与宇宙]为什么发射和回收航天器有档期
comsci
地球的大气层中有一个时空屏蔽层,这个层次会不定时的出现,如果该时空屏蔽层出现,那么将导致外层空间进入的任何物体被摧毁,而从地面发射到太空的飞船也将被摧毁...
所以,航天发射和飞船回收都需要等待这个时空屏蔽层消失之后,再进行
&
- linux下批量替换文件内容
商人shang
linux替换
1、网络上现成的资料
格式: sed -i "s/查找字段/替换字段/g" `grep 查找字段 -rl 路径`
linux sed 批量替换多个文件中的字符串
sed -i "s/oldstring/newstring/g" `grep oldstring -rl yourdir`
例如:替换/home下所有文件中的www.admi
- 网页在线天气预报
oloz
天气预报
网页在线调用天气预报
<%@ page language="java" contentType="text/html; charset=utf-8"
pageEncoding="utf-8"%>
<!DOCTYPE html PUBLIC "-//W3C//DTD HTML 4.01 Transit
- SpringMVC和Struts2比较
杨白白
springMVC
1. 入口
spring mvc的入口是servlet,而struts2是filter(这里要指出,filter和servlet是不同的。以前认为filter是servlet的一种特殊),这样就导致了二者的机制不同,这里就牵涉到servlet和filter的区别了。
参见:http://blog.csdn.net/zs15932616453/article/details/8832343
2
- refuse copy, lazy girl!
小桔子
copy
妹妹坐船头啊啊啊啊!都打算一点点琢磨呢。文字编辑也写了基本功能了。。今天查资料,结果查到了人家写得完完整整的。我清楚的认识到:
1.那是我自己觉得写不出的高度
2.如果直接拿来用,很快就能解决问题
3.然后就是抄咩~~
4.肿么可以这样子,都不想写了今儿个,留着作参考吧!拒绝大抄特抄,慢慢一点点写!
- apache与php整合
aichenglong
php apache web
一 apache web服务器
1 apeche web服务器的安装
1)下载Apache web服务器
2)配置域名(如果需要使用要在DNS上注册)
3)测试安装访问http://localhost/验证是否安装成功
2 apache管理
1)service.msc进行图形化管理
2)命令管理,配
- Maven常用内置变量
AILIKES
maven
Built-in properties
${basedir} represents the directory containing pom.xml
${version} equivalent to ${project.version} (deprecated: ${pom.version})
Pom/Project properties
Al
- java的类和对象
百合不是茶
JAVA面向对象 类 对象
java中的类:
java是面向对象的语言,解决问题的核心就是将问题看成是一个类,使用类来解决
java使用 class 类名 来创建类 ,在Java中类名要求和构造方法,Java的文件名是一样的
创建一个A类:
class A{
}
java中的类:将某两个事物有联系的属性包装在一个类中,再通
- JS控制页面输入框为只读
bijian1013
JavaScript
在WEB应用开发当中,增、删除、改、查功能必不可少,为了减少以后维护的工作量,我们一般都只做一份页面,通过传入的参数控制其是新增、修改或者查看。而修改时需将待修改的信息从后台取到并显示出来,实际上就是查看的过程,唯一的区别是修改时,页面上所有的信息能修改,而查看页面上的信息不能修改。因此完全可以将其合并,但通过前端JS将查看页面的所有信息控制为只读,在信息量非常大时,就比较麻烦。
- AngularJS与服务器交互
bijian1013
JavaScriptAngularJS$http
对于AJAX应用(使用XMLHttpRequests)来说,向服务器发起请求的传统方式是:获取一个XMLHttpRequest对象的引用、发起请求、读取响应、检查状态码,最后处理服务端的响应。整个过程示例如下:
var xmlhttp = new XMLHttpRequest();
xmlhttp.onreadystatechange
- [Maven学习笔记八]Maven常用插件应用
bit1129
maven
常用插件及其用法位于:http://maven.apache.org/plugins/
1. Jetty server plugin
2. Dependency copy plugin
3. Surefire Test plugin
4. Uber jar plugin
1. Jetty Pl
- 【Hive六】Hive用户自定义函数(UDF)
bit1129
自定义函数
1. 什么是Hive UDF
Hive是基于Hadoop中的MapReduce,提供HQL查询的数据仓库。Hive是一个很开放的系统,很多内容都支持用户定制,包括:
文件格式:Text File,Sequence File
内存中的数据格式: Java Integer/String, Hadoop IntWritable/Text
用户提供的 map/reduce 脚本:不管什么
- 杀掉nginx进程后丢失nginx.pid,如何重新启动nginx
ronin47
nginx 重启 pid丢失
nginx进程被意外关闭,使用nginx -s reload重启时报如下错误:nginx: [error] open() “/var/run/nginx.pid” failed (2: No such file or directory)这是因为nginx进程被杀死后pid丢失了,下一次再开启nginx -s reload时无法启动解决办法:nginx -s reload 只是用来告诉运行中的ng
- UI设计中我们为什么需要设计动效
brotherlamp
UIui教程ui视频ui资料ui自学
随着国际大品牌苹果和谷歌的引领,最近越来越多的国内公司开始关注动效设计了,越来越多的团队已经意识到动效在产品用户体验中的重要性了,更多的UI设计师们也开始投身动效设计领域。
但是说到底,我们到底为什么需要动效设计?或者说我们到底需要什么样的动效?做动效设计也有段时间了,于是尝试用一些案例,从产品本身出发来说说我所思考的动效设计。
一、加强体验舒适度
嗯,就是让用户更加爽更加爽的用你的产品。
- Spring中JdbcDaoSupport的DataSource注入问题
bylijinnan
javaspring
参考以下两篇文章:
http://www.mkyong.com/spring/spring-jdbctemplate-jdbcdaosupport-examples/
http://stackoverflow.com/questions/4762229/spring-ldap-invoking-setter-methods-in-beans-configuration
Sprin
- 数据库连接池的工作原理
chicony
数据库连接池
随着信息技术的高速发展与广泛应用,数据库技术在信息技术领域中的位置越来越重要,尤其是网络应用和电子商务的迅速发展,都需要数据库技术支持动 态Web站点的运行,而传统的开发模式是:首先在主程序(如Servlet、Beans)中建立数据库连接;然后进行SQL操作,对数据库中的对象进行查 询、修改和删除等操作;最后断开数据库连接。使用这种开发模式,对
- java 关键字
CrazyMizzz
java
关键字是事先定义的,有特别意义的标识符,有时又叫保留字。对于保留字,用户只能按照系统规定的方式使用,不能自行定义。
Java中的关键字按功能主要可以分为以下几类:
(1)访问修饰符
public,private,protected
p
- Hive中的排序语法
daizj
排序hiveorder byDISTRIBUTE BYsort by
Hive中的排序语法 2014.06.22 ORDER BY
hive中的ORDER BY语句和关系数据库中的sql语法相似。他会对查询结果做全局排序,这意味着所有的数据会传送到一个Reduce任务上,这样会导致在大数量的情况下,花费大量时间。
与数据库中 ORDER BY 的区别在于在hive.mapred.mode = strict模式下,必须指定 limit 否则执行会报错。
- 单态设计模式
dcj3sjt126com
设计模式
单例模式(Singleton)用于为一个类生成一个唯一的对象。最常用的地方是数据库连接。 使用单例模式生成一个对象后,该对象可以被其它众多对象所使用。
<?phpclass Example{ // 保存类实例在此属性中 private static&
- svn locked
dcj3sjt126com
Lock
post-commit hook failed (exit code 1) with output:
svn: E155004: Working copy 'D:\xx\xxx' locked
svn: E200031: sqlite: attempt to write a readonly database
svn: E200031: sqlite: attempt to write a
- ARM寄存器学习
e200702084
数据结构C++cC#F#
无论是学习哪一种处理器,首先需要明确的就是这种处理器的寄存器以及工作模式。
ARM有37个寄存器,其中31个通用寄存器,6个状态寄存器。
1、不分组寄存器(R0-R7)
不分组也就是说说,在所有的处理器模式下指的都时同一物理寄存器。在异常中断造成处理器模式切换时,由于不同的处理器模式使用一个名字相同的物理寄存器,就是
- 常用编码资料
gengzg
编码
List<UserInfo> list=GetUserS.GetUserList(11);
String json=JSON.toJSONString(list);
HashMap<Object,Object> hs=new HashMap<Object, Object>();
for(int i=0;i<10;i++)
{
- 进程 vs. 线程
hongtoushizi
线程linux进程
我们介绍了多进程和多线程,这是实现多任务最常用的两种方式。现在,我们来讨论一下这两种方式的优缺点。
首先,要实现多任务,通常我们会设计Master-Worker模式,Master负责分配任务,Worker负责执行任务,因此,多任务环境下,通常是一个Master,多个Worker。
如果用多进程实现Master-Worker,主进程就是Master,其他进程就是Worker。
如果用多线程实现
- Linux定时Job:crontab -e 与 /etc/crontab 的区别
Josh_Persistence
linuxcrontab
一、linux中的crotab中的指定的时间只有5个部分:* * * * *
分别表示:分钟,小时,日,月,星期,具体说来:
第一段 代表分钟 0—59
第二段 代表小时 0—23
第三段 代表日期 1—31
第四段 代表月份 1—12
第五段 代表星期几,0代表星期日 0—6
如:
*/1 * * * * 每分钟执行一次。
*
- KMP算法详解
hm4123660
数据结构C++算法字符串KMP
字符串模式匹配我们相信大家都有遇过,然而我们也习惯用简单匹配法(即Brute-Force算法),其基本思路就是一个个逐一对比下去,这也是我们大家熟知的方法,然而这种算法的效率并不高,但利于理解。
假设主串s="ababcabcacbab",模式串为t="
- 枚举类型的单例模式
zhb8015
单例模式
E.编写一个包含单个元素的枚举类型[极推荐]。代码如下:
public enum MaYun {himself; //定义一个枚举的元素,就代表MaYun的一个实例private String anotherField;MaYun() {//MaYun诞生要做的事情//这个方法也可以去掉。将构造时候需要做的事情放在instance赋值的时候:/** himself = MaYun() {*
- Kafka+Storm+HDFS
ssydxa219
storm
cd /myhome/usr/stormbin/storm nimbus &bin/storm supervisor &bin/storm ui &Kafka+Storm+HDFS整合实践kafka_2.9.2-0.8.1.1.tgzapache-storm-0.9.2-incubating.tar.gzKafka安装配置我们使用3台机器搭建Kafk
- Java获取本地服务器的IP
中华好儿孙
javaWeb获取服务器ip地址
System.out.println("getRequestURL:"+request.getRequestURL());
System.out.println("getLocalAddr:"+request.getLocalAddr());
System.out.println("getLocalPort:&quo
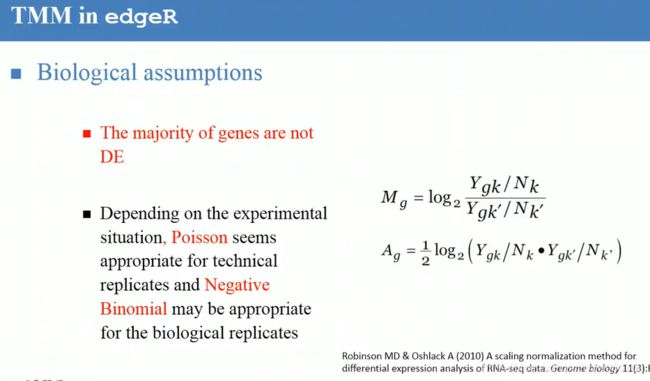
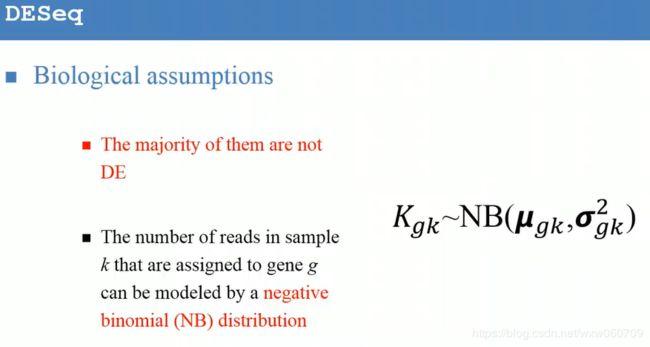
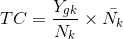 , Total count(TC) (具体可以参考阅读paper list中文献)
, Total count(TC) (具体可以参考阅读paper list中文献)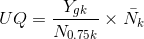 , Upper Quartile(UQ)
, Upper Quartile(UQ)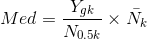 , Median(Med)
, Median(Med)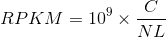 , Reads Per Kilobase per Million(RPKM), C: the number of reads mapped onto the gene's exons; N:total number of reads in the experiment; L: the sum of the exons in base pairs.
, Reads Per Kilobase per Million(RPKM), C: the number of reads mapped onto the gene's exons; N:total number of reads in the experiment; L: the sum of the exons in base pairs.