UACANet: Uncertainty Augmented Context Attention for Polyp Segmentation代码补充
上一篇看了文章创新点的代码,现在看一下train文件等其余的文件。
看主函数:
import os
import torch
import argparse
import tqdm
import sys
import cv2
import torch.nn as nn
import torch.distributed as dist
from torch.optim import Adam, SGD
from torch.cuda.amp import GradScaler, autocast
filepath = os.path.split(__file__)[0]
repopath = os.path.split(filepath)[0]
sys.path.append(repopath)
from utils.dataloader import *
from lib.optim import *
from lib import *
def _args():
parser = argparse.ArgumentParser()
parser.add_argument('--config', type=str, default='configs/UACANet-L.yaml')
parser.add_argument('--local_rank', type=int, default=-1)
parser.add_argument('--verbose', action='store_true', default=False)
parser.add_argument('--debug', action='store_true', default=False)
return parser.parse_args()
def train(opt, args):
device_ids = os.environ["CUDA_VISIBLE_DEVICES"].split(',')
device_num = len(device_ids)
train_dataset = eval(opt.Train.Dataset.type)(
root=opt.Train.Dataset.root, transform_list=opt.Train.Dataset.transform_list)
if device_num > 1:
torch.cuda.set_device(args.local_rank)
dist.init_process_group(backend='nccl')
train_sampler = torch.utils.data.distributed.DistributedSampler(
train_dataset, shuffle=True)
else:
train_sampler = None
train_loader = data.DataLoader(dataset=train_dataset,
batch_size=opt.Train.Dataloader.batch_size,
shuffle=train_sampler is None,
sampler=train_sampler,
num_workers=opt.Train.Dataloader.num_workers,
pin_memory=opt.Train.Dataloader.pin_memory,
drop_last=True)
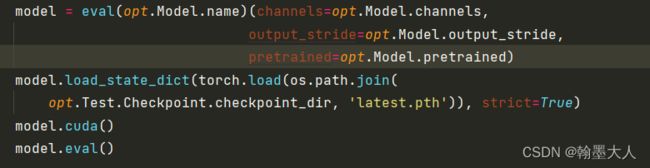
model = eval(opt.Model.name)(channels=opt.Model.channels,
output_stride=opt.Model.output_stride,
pretrained=opt.Model.pretrained)
if device_num > 1:
model = nn.SyncBatchNorm.convert_sync_batchnorm(model)
model = model.cuda()
model = nn.parallel.DistributedDataParallel(model, device_ids=[
args.local_rank], output_device=args.local_rank, find_unused_parameters=True)
else:
model = model.cuda()
backbone_params = nn.ParameterList()
decoder_params = nn.ParameterList()
for name, param in model.named_parameters():
if 'backbone' in name:
if 'backbone.layer' in name:
backbone_params.append(param)
else:
pass
else:
decoder_params.append(param)
params_list = [{'params': backbone_params}, {
'params': decoder_params, 'lr': opt.Train.Optimizer.lr * 10}]
optimizer = eval(opt.Train.Optimizer.type)(
params_list, opt.Train.Optimizer.lr, weight_decay=opt.Train.Optimizer.weight_decay)
if opt.Train.Optimizer.mixed_precision is True:
scaler = GradScaler()
else:
scaler = None
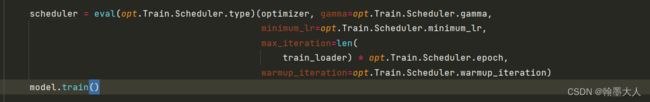
scheduler = eval(opt.Train.Scheduler.type)(optimizer, gamma=opt.Train.Scheduler.gamma,
minimum_lr=opt.Train.Scheduler.minimum_lr,
max_iteration=len(
train_loader) * opt.Train.Scheduler.epoch,
warmup_iteration=opt.Train.Scheduler.warmup_iteration)
model.train()
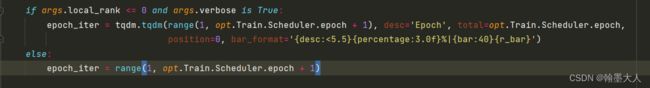
if args.local_rank <= 0 and args.verbose is True:
epoch_iter = tqdm.tqdm(range(1, opt.Train.Scheduler.epoch + 1), desc='Epoch', total=opt.Train.Scheduler.epoch,
position=0, bar_format='{desc:<5.5}{percentage:3.0f}%|{bar:40}{r_bar}')
else:
epoch_iter = range(1, opt.Train.Scheduler.epoch + 1)
for epoch in epoch_iter:
if args.local_rank <= 0 and args.verbose is True:
step_iter = tqdm.tqdm(enumerate(train_loader, start=1), desc='Iter', total=len(
train_loader), position=1, leave=False, bar_format='{desc:<5.5}{percentage:3.0f}%|{bar:40}{r_bar}')
if device_num > 1:
train_sampler.set_epoch(epoch)
else:
step_iter = enumerate(train_loader, start=1)
for i, sample in step_iter:
optimizer.zero_grad()
if opt.Train.Optimizer.mixed_precision is True:
with autocast():
sample = to_cuda(sample)
out = model(sample)
scaler.scale(out['loss']).backward()
scaler.step(optimizer)
scaler.update()
scheduler.step()
else:
sample = to_cuda(sample)
out = model(sample)
out['loss'].backward()
optimizer.step()
scheduler.step()
if args.local_rank <= 0 and args.verbose is True:
step_iter.set_postfix({'loss': out['loss'].item()})
if args.local_rank <= 0:
os.makedirs(opt.Train.Checkpoint.checkpoint_dir, exist_ok=True)
os.makedirs(os.path.join(
opt.Train.Checkpoint.checkpoint_dir, 'debug'), exist_ok=True)
if epoch % opt.Train.Checkpoint.checkpoint_epoch == 0:
torch.save(model.module.state_dict() if device_num > 1 else model.state_dict(
), os.path.join(opt.Train.Checkpoint.checkpoint_dir, 'latest.pth'))
if args.debug is True:
debout = debug_tile(out)
cv2.imwrite(os.path.join(
opt.Train.Checkpoint.checkpoint_dir, 'debug', str(epoch) + '.png'), debout)
if args.local_rank <= 0:
torch.save(model.module.state_dict() if device_num > 1 else model.state_dict(
), os.path.join(opt.Train.Checkpoint.checkpoint_dir, 'latest.pth'))
if __name__ == '__main__':
args = _args()
opt = load_config(args.config)
train(opt, args)
args = 模型的配置
opt= load_config(args.config)=载入配置config配置。
首先打开配置的地址,载入yaml文件,yaml.FullLoader意思为加载完整的YAML语言。避免任意代码执行。这是当前(PyYAML5.1)默认加载器调用。
def load_config(config_dir):
return ed(yaml.load(open(config_dir), yaml.FullLoader))
yaml文件内部:
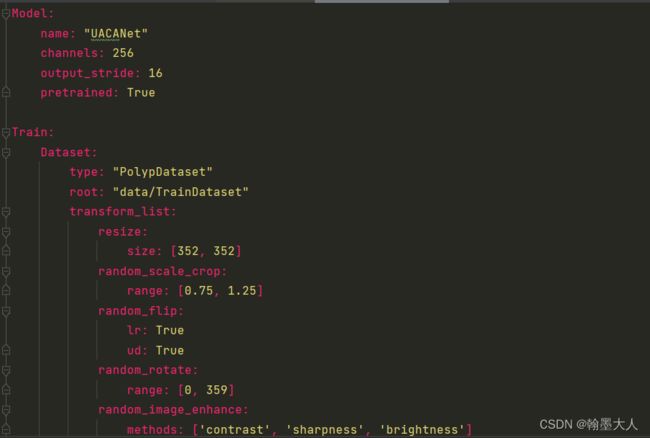
yaml文件内容类型:
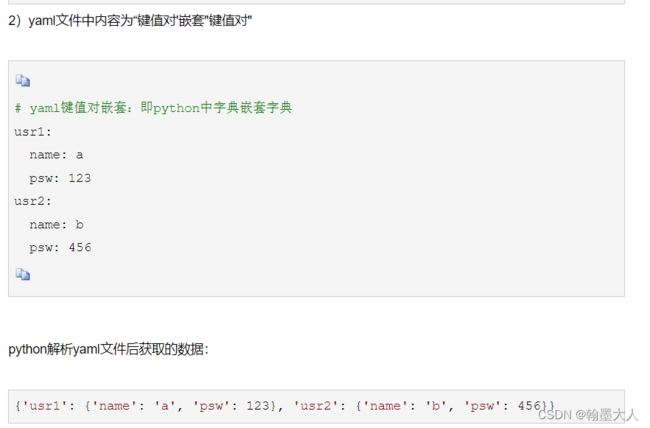
那么yaml文件返回的格式也类似于{‘model’:{‘name’:‘UACANet’,…} , ‘train’:{‘dataset’:{‘type’:‘polydataset’},…}。前面的ed代表是easydict,这样可以更方便的的获取字典的属性。
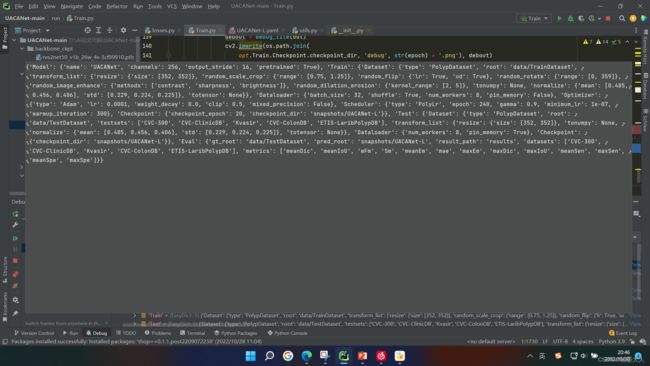
然后我们进入train函数:
1:首先获得cuda的id和个数。
2:然后我们获得yaml文件下,Train.dataset.type=“PolypDataset”。然后是eval函数:将字符串转为python语句(就是去掉“”)然后执行转化后的语句。第二个括号是PolypDataset的根目录:data/TestDataset,接着是要对图片执行的变换,包括resize等属性。
相当于train_dataset = PolypDataset(root=‘’,…)。
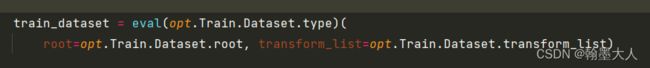
3:如果gpu数量多于1个就可以使用分布式训练。
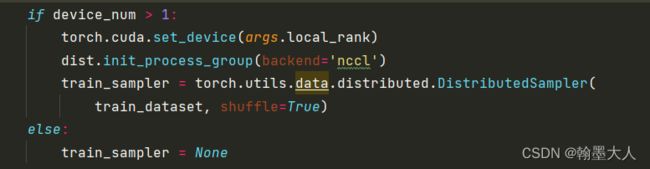
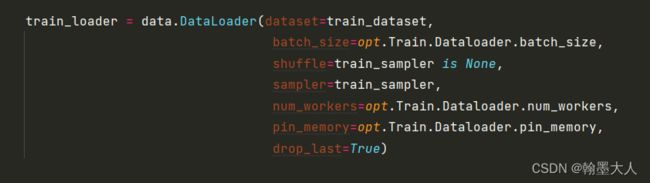
4:然后模型载入数据指定数据集,batchsize,numworkers等。
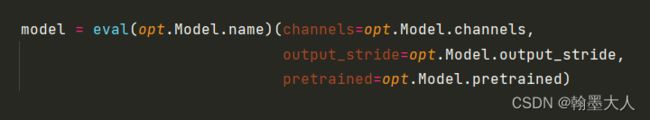
5:然后获得模型的名字name: “UACANet”,通道256等参数。
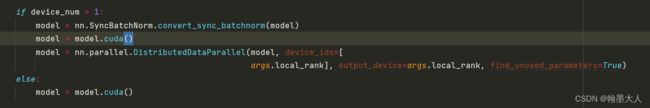
6:将模型送入到cuda中,分别为多gpu和单gpu‘的方法。’
7:模型的backbone定义两个空parameterlist。注意在训练时候是不断更新的。ParameterList输入一定是一个Parameter的List,其他类型会报错,在注册时候就会提示元素不是Parameter类型。
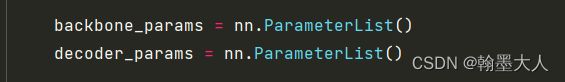
8:我们遍历model的参数,将含有backbone.layer对应的参数添加到backbone_params,(我这里打印的参数没有backbone.layer可能是没有载入权重),将剩余的载入decoder_params中。
打印一下name:
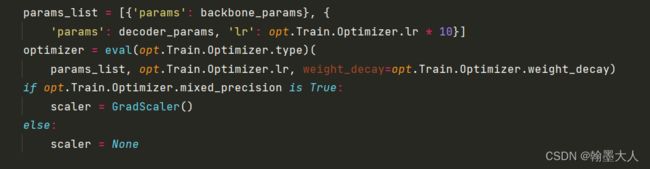
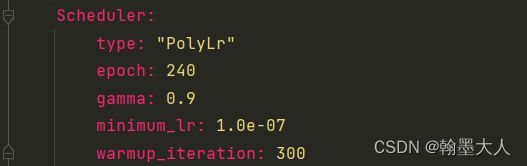
9:现在的param_liat包含了param,param,lr三个字典。优化器参数: Optimizer:
type: “Adam”
lr: 1.0e-04
weight_decay: 0.0
clip: 0.5
mixed_precision: False
10:迭代更新策略:
11:更新epoch的次数,tqdm是显示进度条。
12:接着就是在一个epoch里面每次取batch个数据进行处理。enumerate将trainloader组合成一个索引序列,同时给出数据。
然后遍历step_liter。优化器清零,将sample输入到cuda中,然后在输入到模型中,获得一个输出。
for epoch in epoch_iter:
if args.local_rank <= 0 and args.verbose is True:
step_iter = tqdm.tqdm(enumerate(train_loader, start=1), desc='Iter', total=len(
train_loader), position=1, leave=False, bar_format='{desc:<5.5}{percentage:3.0f}%|{bar:40}{r_bar}')
if device_num > 1:
train_sampler.set_epoch(epoch)
else:
step_iter = enumerate(train_loader, start=1)
for i, sample in step_iter:
optimizer.zero_grad()
if opt.Train.Optimizer.mixed_precision is True:
with autocast():
sample = to_cuda(sample)
out = model(sample)
scaler.scale(out['loss']).backward()
scaler.step(optimizer)
scaler.update()
scheduler.step()
else:
sample = to_cuda(sample)
out = model(sample)
out['loss'].backward()
optimizer.step()
scheduler.step()
**注意:在model中会计算模型的loss。**将中间生成的输出与原始gt进行损失计算。
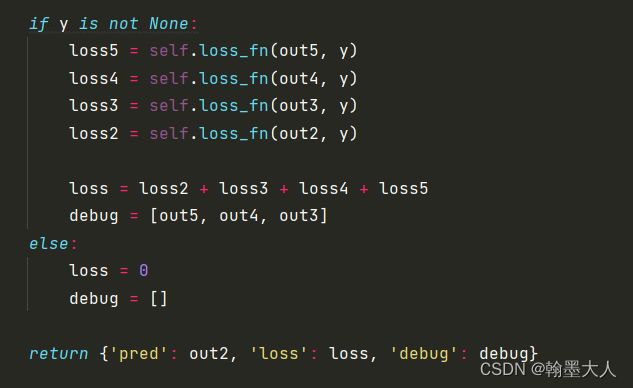
我们根据loss跳到bceloss函数中。pred是预测输出,mask是gt。
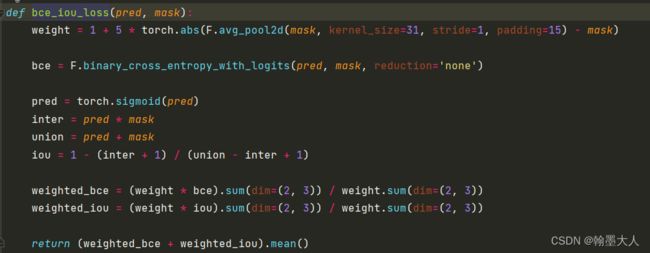
首先mask经过下采样不变,减去本身等于0,那么weight=1。bce直接计算,有现成的函数。
将输出经过一个sigmoid函数,计算inter为预测值和真实值相乘,union为预测值和真实值相加,那么iou_loss= 1 - (inter + 1) / (union - inter + 1),分子分母都加一是防止相除为0.
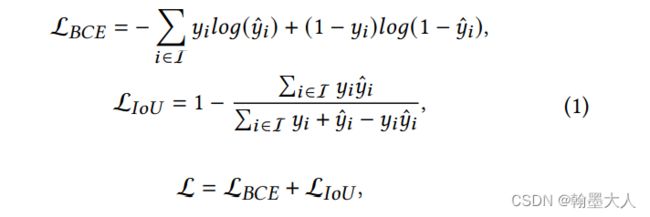
然后经过权重后的两个损失相加求均值。
然后我们损失反向更新,优化器和迭代策略更新。
接着开辟目录"snapshots/UACANet-L",在"snapshots/UACANet-L"目录下开辟新的目录debug。
如果当前epoch除以20可以整除,保存模型的参数,和最后一代。
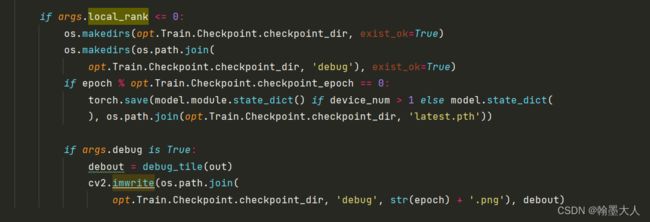
-------------------------------------------------------------------整个train文件就结束了---------------------------------------------------------------------------------------
test文件是测试数据集的预测图,eval文件是评估产生的预测图。
首先看test文件:
def _args():
parser = argparse.ArgumentParser()
parser.add_argument('--config', type=str, default='configs/UACANet-L.yaml')
parser.add_argument('--verbose', action='store_true', default=False)
return parser.parse_args()
def test(opt, args):
model = eval(opt.Model.name)(channels=opt.Model.channels,
output_stride=opt.Model.output_stride,
pretrained=opt.Model.pretrained)
model.load_state_dict(torch.load(os.path.join(
opt.Test.Checkpoint.checkpoint_dir, 'latest.pth')), strict=True)
model.cuda()
model.eval()
if args.verbose is True:
testsets = tqdm.tqdm(opt.Test.Dataset.testsets, desc='Total TestSet', total=len(
opt.Test.Dataset.testsets), position=0, bar_format='{desc:<30}{percentage:3.0f}%|{bar:50}{r_bar}')
else:
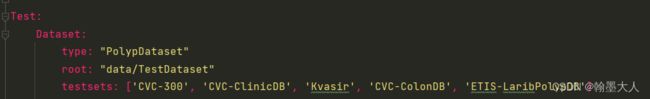
testsets = opt.Test.Dataset.testsets
for testset in testsets:
data_path = os.path.join(opt.Test.Dataset.root, testset)
save_path = os.path.join(opt.Test.Checkpoint.checkpoint_dir, testset)
os.makedirs(save_path, exist_ok=True)
test_dataset = eval(opt.Test.Dataset.type)(root=data_path, transform_list=opt.Test.Dataset.transform_list)
test_loader = data.DataLoader(dataset=test_dataset,
batch_size=1,
num_workers=opt.Test.Dataloader.num_workers,
pin_memory=opt.Test.Dataloader.pin_memory)
if args.verbose is True:
samples = tqdm.tqdm(test_loader, desc=testset + ' - Test', total=len(test_loader),
position=1, leave=False, bar_format='{desc:<30}{percentage:3.0f}%|{bar:50}{r_bar}')
else:
samples = test_loader
for sample in samples:
sample = to_cuda(sample)
out = model(sample)
out['pred'] = F.interpolate(
out['pred'], sample['shape'], mode='bilinear', align_corners=True)
out['pred'] = out['pred'].data.cpu()
out['pred'] = torch.sigmoid(out['pred'])
out['pred'] = out['pred'].numpy().squeeze()
out['pred'] = (out['pred'] - out['pred'].min()) / \
(out['pred'].max() - out['pred'].min() + 1e-8)
Image.fromarray(((out['pred'] > .5) * 255).astype(np.uint8)
).save(os.path.join(save_path, sample['name'][0]))
if __name__ == "__main__":
args = _args()
opt = load_config(args.config)
test(opt, args)
1:和train文件一样,首先载入配置。
2:获得模型的名字和属性,载入最新一代的权重,输送到cuda中,将模型设置为评估模式。
3:测试数据集不止一个,遍历第一个数据集,首先获得数据集路径,然后指定保存路径。开辟保存路径。
for testset in testsets:
data_path = os.path.join(opt.Test.Dataset.root, testset)
save_path = os.path.join(opt.Test.Checkpoint.checkpoint_dir, testset)
os.makedirs(save_path, exist_ok=True)
test_dataset = eval(opt.Test.Dataset.type)(root=data_path, transform_list=opt.Test.Dataset.transform_list)
test_loader = data.DataLoader(dataset=test_dataset,
batch_size=1,
num_workers=opt.Test.Dataloader.num_workers,
pin_memory=opt.Test.Dataloader.pin_memory)
if args.verbose is True:
samples = tqdm.tqdm(test_loader, desc=testset + ' - Test', total=len(test_loader),
position=1, leave=False, bar_format='{desc:<30}{percentage:3.0f}%|{bar:50}{r_bar}')
else:
samples = test_loader
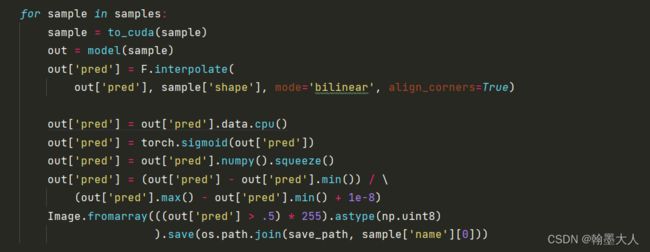
for sample in samples:
sample = to_cuda(sample)
out = model(sample)
out['pred'] = F.interpolate(
out['pred'], sample['shape'], mode='bilinear', align_corners=True)
out['pred'] = out['pred'].data.cpu()
out['pred'] = torch.sigmoid(out['pred'])
out['pred'] = out['pred'].numpy().squeeze()
out['pred'] = (out['pred'] - out['pred'].min()) / \
(out['pred'].max() - out['pred'].min() + 1e-8)
Image.fromarray(((out['pred'] > .5) * 255).astype(np.uint8)
).save(os.path.join(save_path, sample['name'][0]))
指定测试数据集,加载数据集的图片,每一加载一张,sample=加载的数据。
遍历sample,将sample输入到cuda中,然后输入到模型中,将输出的预测图采样到原图像大小。
将输出值放到gpu上,经过sigmoid函数,再压缩第一个维度,进行归一化。
image.formarray将array转换到图片,保存在save_path中。
-----------------------------------------------------------------------test结束---------------------------------------------------------------------------------------
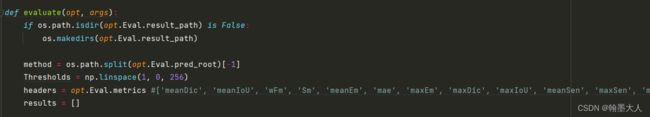
def evaluate(opt, args):
if os.path.isdir(opt.Eval.result_path) is False:
os.makedirs(opt.Eval.result_path)
method = os.path.split(opt.Eval.pred_root)[-1]
Thresholds = np.linspace(1, 0, 256)
headers = opt.Eval.metrics #['meanDic', 'meanIoU', 'wFm', 'Sm', 'meanEm', 'mae', 'maxEm', 'maxDic', 'maxIoU', 'meanSen', 'maxSen', 'meanSpe', 'maxSpe']
results = []
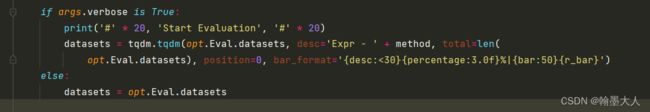
if args.verbose is True:
print('#' * 20, 'Start Evaluation', '#' * 20)
datasets = tqdm.tqdm(opt.Eval.datasets, desc='Expr - ' + method, total=len(
opt.Eval.datasets), position=0, bar_format='{desc:<30}{percentage:3.0f}%|{bar:50}{r_bar}')
else:
datasets = opt.Eval.datasets
for dataset in datasets:
pred_root = os.path.join(opt.Eval.pred_root, dataset)
gt_root = os.path.join(opt.Eval.gt_root, dataset, 'masks')
preds = os.listdir(pred_root)
gts = os.listdir(gt_root)
preds.sort()
gts.sort()
threshold_Fmeasure = np.zeros((len(preds), len(Thresholds)))
threshold_Emeasure = np.zeros((len(preds), len(Thresholds)))
threshold_IoU = np.zeros((len(preds), len(Thresholds)))
# threshold_Precision = np.zeros((len(preds), len(Thresholds)))
# threshold_Recall = np.zeros((len(preds), len(Thresholds)))
threshold_Sensitivity = np.zeros((len(preds), len(Thresholds)))
threshold_Specificity = np.zeros((len(preds), len(Thresholds)))
threshold_Dice = np.zeros((len(preds), len(Thresholds)))
Smeasure = np.zeros(len(preds))
wFmeasure = np.zeros(len(preds))
MAE = np.zeros(len(preds))
if args.verbose is True:
samples = tqdm.tqdm(enumerate(zip(preds, gts)), desc=dataset + ' - Evaluation', total=len(
preds), position=1, leave=False, bar_format='{desc:<30}{percentage:3.0f}%|{bar:50}{r_bar}')
else:
samples = enumerate(zip(preds, gts))
for i, sample in samples:
pred, gt = sample
assert os.path.splitext(pred)[0] == os.path.splitext(gt)[0]
pred_mask = np.array(Image.open(os.path.join(pred_root, pred)))
gt_mask = np.array(Image.open(os.path.join(gt_root, gt)))
if len(pred_mask.shape) != 2:
pred_mask = pred_mask[:, :, 0]
if len(gt_mask.shape) != 2:
gt_mask = gt_mask[:, :, 0]
assert pred_mask.shape == gt_mask.shape
gt_mask = gt_mask.astype(np.float64) / 255
gt_mask = (gt_mask > 0.5).astype(np.float64)
pred_mask = pred_mask.astype(np.float64) / 255
Smeasure[i] = StructureMeasure(pred_mask, gt_mask)
wFmeasure[i] = original_WFb(pred_mask, gt_mask)
MAE[i] = np.mean(np.abs(gt_mask - pred_mask))
threshold_E = np.zeros(len(Thresholds))
threshold_F = np.zeros(len(Thresholds))
threshold_Pr = np.zeros(len(Thresholds))
threshold_Rec = np.zeros(len(Thresholds))
threshold_Iou = np.zeros(len(Thresholds))
threshold_Spe = np.zeros(len(Thresholds))
threshold_Dic = np.zeros(len(Thresholds))
for j, threshold in enumerate(Thresholds):
threshold_Pr[j], threshold_Rec[j], threshold_Spe[j], threshold_Dic[j], threshold_F[j], threshold_Iou[j] = Fmeasure_calu(pred_mask, gt_mask, threshold)
Bi_pred = np.zeros_like(pred_mask)
Bi_pred[pred_mask >= threshold] = 1
threshold_E[j] = EnhancedMeasure(Bi_pred, gt_mask)
threshold_Emeasure[i, :] = threshold_E
threshold_Fmeasure[i, :] = threshold_F
threshold_Sensitivity[i, :] = threshold_Rec
threshold_Specificity[i, :] = threshold_Spe
threshold_Dice[i, :] = threshold_Dic
threshold_IoU[i, :] = threshold_Iou
result = []
mae = np.mean(MAE)
Sm = np.mean(Smeasure)
wFm = np.mean(wFmeasure)
column_E = np.mean(threshold_Emeasure, axis=0)
meanEm = np.mean(column_E)
maxEm = np.max(column_E)
column_Sen = np.mean(threshold_Sensitivity, axis=0)
meanSen = np.mean(column_Sen)
maxSen = np.max(column_Sen)
column_Spe = np.mean(threshold_Specificity, axis=0)
meanSpe = np.mean(column_Spe)
maxSpe = np.max(column_Spe)
column_Dic = np.mean(threshold_Dice, axis=0)
meanDic = np.mean(column_Dic)
maxDic = np.max(column_Dic)
column_IoU = np.mean(threshold_IoU, axis=0)
meanIoU = np.mean(column_IoU)
maxIoU = np.max(column_IoU)
# result.extend([meanDic, meanIoU, wFm, Sm, meanEm, mae, maxEm, maxDic, maxIoU, meanSen, maxSen, meanSpe, maxSpe])
# results.append([dataset, *result])
out = []
for metric in opt.Eval.metrics:
out.append(eval(metric))
result.extend(out)
results.append([dataset, *result])
csv = os.path.join(opt.Eval.result_path, 'result_' + dataset + '.csv')
if os.path.isfile(csv) is True:
csv = open(csv, 'a')
else:
csv = open(csv, 'w')
csv.write(', '.join(['method', *headers]) + '\n')
out_str = method + ','
for metric in result:
out_str += '{:.4f}'.format(metric) + ','
out_str += '\n'
csv.write(out_str)
csv.close()
tab = tabulate(results, headers=['dataset', *headers], floatfmt=".3f")
if args.verbose is True:
print(tab)
print("#"*20, "End Evaluation", "#"*20)
return tab
if __name__ == "__main__":
args = _args()
opt = load_config(args.config)
evaluate(opt, args)
1:如果结果路径不存在,重新创建一个。
2:将pred_root划分出来,split函数。然后在1-0之间等间隔划分256个数作为阈值(递减)。header为评价指标。
3:指定评估的dataset。
4:遍历dataset,获得预测的路径和gt路径。打开预测路径,打开gt路径。对路径里面的图片进行排序。添加链接描述
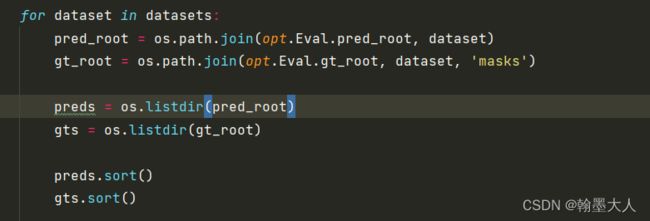
5:生成len(preds)行, len(Thresholds)列的0矩阵。生成一行len(preds)列的0列表。
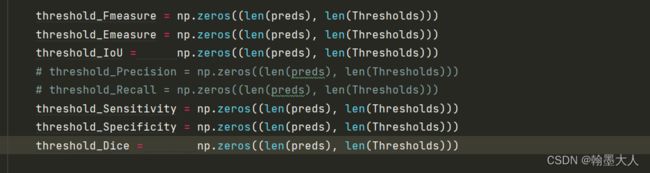
6:遍历samples,sample为pred和gt的一一配对。打开pred和gt,并转换为array格式。
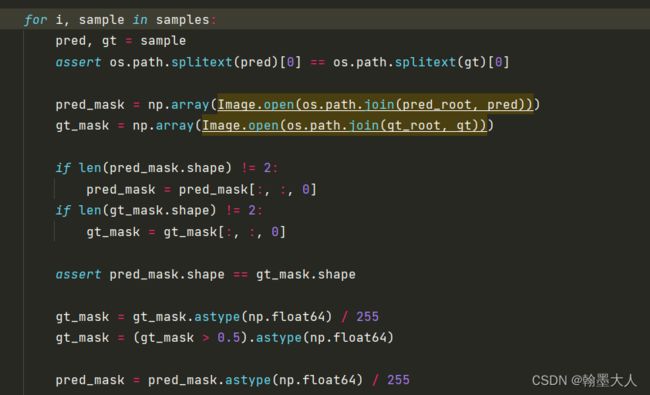
取pred的三维:
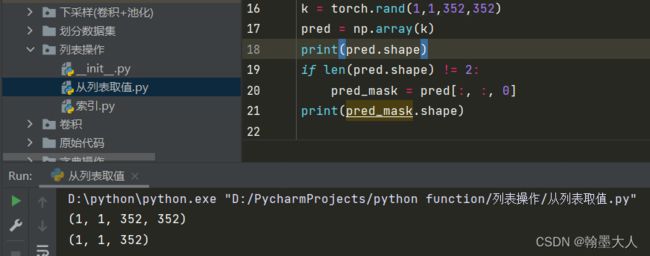
7:取gt和pred:
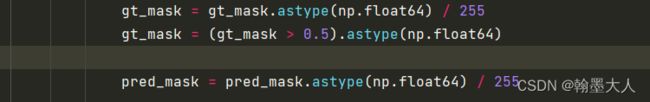
8:计算pred和gt之间的损失,做后用table格式表现出来。