wgcna 实战 jimmy 全流程 全代码
input
getwd()#“G:/linux study/hsp70_human/ref/my_WGCNA-master”
> table(datTraits$subtype)
Basal Claudin-low Luminal Non-malignant unknown
14 6 27 5 4
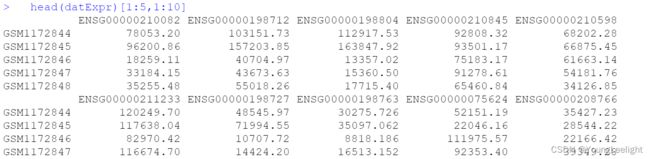
fpkm[1:4,1:4]
head(datTraits)
table(datTraits$subtype)
RNAseq_voom <- fpkm
## 因为WGCNA针对的是基因进行聚类,而一般我们的聚类是针对样本用hclust即可,所以这个时候需要转置
WGCNA_matrix = t(RNAseq_voom[order(apply(RNAseq_voom,1,mad), decreasing = T)[1:5000],])
datExpr0 <- WGCNA_matrix ## top 5000 mad genes
datExpr <- datExpr0
head(datExpr)[1:5,1:10]
dynamicColors = labels2colors(dynamicMods)
table(dynamicColors)
# Plot the dendrogram and colors underneath
#sizeGrWindow(8,6)
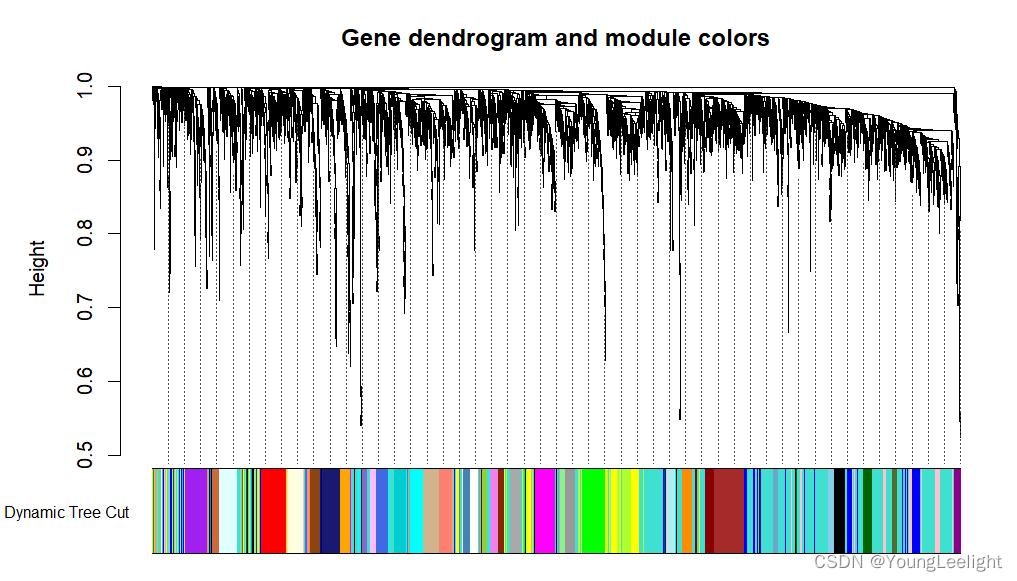
plotDendroAndColors(geneTree, dynamicColors, "Dynamic Tree Cut",
dendroLabels = FALSE, hang = 0.03,
addGuide = TRUE, guideHang = 0.05,
main = "Gene dendrogram and module colors")
## 这一步主要是针对于连续变量,如果是分类变量,需要转换成连续变量方可使用
table(datTraits$subtype)
if(T){
nGenes = ncol(datExpr)
nSamples = nrow(datExpr)
design=model.matrix(~0+ datTraits$subtype)
colnames(design)=levels(datTraits$subtype)
moduleColors <- labels2colors(net$colors)
# Recalculate MEs with color labels
MEs0 = moduleEigengenes(datExpr, moduleColors)$eigengenes
MEs = orderMEs(MEs0); ##不同颜色的模块的ME值矩 (样本vs模块)
moduleTraitCor = cor(MEs, design , use = "p");
moduleTraitPvalue = corPvalueStudent(moduleTraitCor, nSamples)
sizeGrWindow(10,6)
# Will display correlations and their p-values
textMatrix = paste(signif(moduleTraitCor, 2), "\n(",
signif(moduleTraitPvalue, 1), ")", sep = "");
dim(textMatrix) = dim(moduleTraitCor)
png("step5-Module-trait-relationships.png",width = 800,height = 1200,res = 120)
par(mar = c(6, 8.5, 3, 3));
# Display the correlation values within a heatmap plot
labeledHeatmap(Matrix = moduleTraitCor,
xLabels = colnames(design),
yLabels = names(MEs),
ySymbols = names(MEs),
colorLabels = FALSE,
colors = greenWhiteRed(50),
textMatrix = textMatrix,
setStdMargins = FALSE,
cex.text = 0.5,
zlim = c(-1,1),
main = paste("Module-trait relationships"))
dev.off()
# 除了上面的热图展现形状与基因模块的相关性外
# 还可以是条形图,但是只能是指定某个形状
# 或者自己循环一下批量出图。
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
y=Luminal
GS1=as.numeric(cor(y,datExpr, use="p"))
GeneSignificance=abs(GS1)
# Next module significance is defined as average gene significance.
ModuleSignificance=tapply(GeneSignificance,
moduleColors, mean, na.rm=T)
sizeGrWindow(8,7)
par(mfrow = c(1,1))
# 如果模块太多,下面的展示就不友好
# 不过,我们可以自定义出图。
plotModuleSignificance(GeneSignificance,moduleColors)
}
## step 6 (第二重要的):感兴趣性状的模块的具体基因分析
# 查看第五步出图:step5-Module-trait-relationships.png
# 发现跟 Luminal 亚型 最相关的是 brown 模块
# 所以接下来就分析这两个
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
module = "brown"
if(T){
# names (colors) of the modules
modNames = substring(names(MEs), 3)
geneModuleMembership = as.data.frame(cor(datExpr, MEs, use = "p"));
## 算出每个模块跟基因的皮尔森相关系数矩
## MEs是每个模块在每个样本里面的
## datExpr是每个基因在每个样本的表达量
MMPvalue = as.data.frame(corPvalueStudent(as.matrix(geneModuleMembership), nSamples));
names(geneModuleMembership) = paste("MM", modNames, sep="");
names(MMPvalue) = paste("p.MM", modNames, sep="");
geneModuleMembership[1:4,1:4]
## 只有连续型性状才能只有计算
## 这里把是否属 Luminal 表型这个变量0,1进行数值化
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
geneTraitSignificance = as.data.frame(cor(datExpr, Luminal, use = "p"));
GSPvalue = as.data.frame(corPvalueStudent(as.matrix(geneTraitSignificance), nSamples));
names(geneTraitSignificance) = paste("GS.", names(Luminal), sep="");
names(GSPvalue) = paste("p.GS.", names(Luminal), sep="");
module = "brown"
column = match(module, modNames);
moduleGenes = moduleColors==module;
png("step6-Module_membership-gene_significance.png",width = 800,height = 600)
#sizeGrWindow(7, 7);
par(mfrow = c(1,1));
verboseScatterplot(abs(geneModuleMembership[moduleGenes, column]),
abs(geneTraitSignificance[moduleGenes, 1]),
xlab = paste("Module Membership in", module, "module"),
ylab = "Gene significance for Luminal",
main = paste("Module membership vs. gene significance\n"),
cex.main = 1.2, cex.lab = 1.2, cex.axis = 1.2, col = module)
dev.off()
}
getwd()#"G:/linux study/hsp70_human/ref/my_WGCNA-master"
rm(list = ls())
options(stringsAsFactors = F)
if(1==1){
# 切换工作目录如果有必要的话
# setwd('WGCNA/')
# 56 breast cancer cell lines were profiled to identify patterns of gene expression associated with subtype and response to therapeutic compounds.
# 这个时候需要制作表达矩阵,每个实例都不一样,代码需要灵活调整
# 我已经制作好了 GSE48213-wgcna-input.RData ,大家直接运行后面的WGCNA代码即可。
if(F){
## https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48213
#wget -c ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE48nnn/GSE48213/suppl/GSE48213_RAW.tar
#tar -xf GSE48213_RAW.tar
#gzip -d *.gz
## 首先在GSE48213_RAW目录里面生成tmp.txt文件,使用shell脚本
# awk '{print FILENAME"\t"$0}' GSM*.txt |grep -v EnsEMBL_Gene_ID >tmp.txt
# 其实也可以直接使用R来读取GSE48213_RAW.tar里面的gz文件,这里就不演示了
# 可以参考:https://mp.weixin.qq.com/s/OLc9QmfN0YcT548VAYgOPA 里面的教程
## 然后把tmp.txt导入R语言里面用reshape2处理即可
# 这个 tmp.txt 文件应该是100M左右大小哦。
}
a=read.table('GSE48213_RAW/tmp.txt',sep = '\t',stringsAsFactors = F)
library(reshape2)
fpkm <- dcast(a,formula = V2~V1)
rownames(fpkm)=fpkm[,1]
fpkm=fpkm[,-1]
colnames(fpkm)=sapply(colnames(fpkm),function(x) strsplit(x,"_")[[1]][1])
library(GEOquery)
a=getGEO('GSE48213')
metadata=pData(a[[1]])[,c(2,10,12)]
datTraits = data.frame(gsm=metadata[,1],
cellline=trimws(sapply(as.character(metadata$characteristics_ch1),function(x) strsplit(x,":")[[1]][2])),
subtype=trimws(sapply(as.character(metadata$characteristics_ch1.2),function(x) strsplit(x,":")[[1]][2]))
)
save(fpkm,datTraits,file = 'GSE48213-wgcna-input.RData')
}
load('GSE48213-wgcna-input.RData')
library(WGCNA)
## step 1 :
if(T){
fpkm[1:4,1:4]
head(datTraits)
table(datTraits$subtype)
RNAseq_voom <- fpkm
## 因为WGCNA针对的是基因进行聚类,而一般我们的聚类是针对样本用hclust即可,所以这个时候需要转置
WGCNA_matrix = t(RNAseq_voom[order(apply(RNAseq_voom,1,mad), decreasing = T)[1:5000],])
datExpr0 <- WGCNA_matrix ## top 5000 mad genes
datExpr <- datExpr0
head(datExpr)[1:5,1:10]
## 下面主要是为了防止临床表型与样本名字对不上
identical(rownames(datExpr),rownames(datTraits))
sampleNames = rownames(datExpr);
traitRows = match(sampleNames, datTraits$gsm)
rownames(datTraits) = datTraits[traitRows, 1]
}
## step 2
datExpr[1:4,1:4]
if(T){
powers = c(c(1:10), seq(from = 12, to=20, by=2))
# Call the network topology analysis function
sft = pickSoftThreshold(datExpr, powerVector = powers, verbose = 5)
#设置网络构建参数选择范围,计算无尺度分布拓扑矩阵
getwd()
#利用sft 画图找软阈值
if(1==1){
png("step2-beta-value.png",width = 800,height = 600)
# Plot the results:
##sizeGrWindow(9, 5)
par(mfrow = c(1,2));
cex1 = 0.9;
# Scale-free topology fit index as a function of the soft-thresholding power
plot(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
xlab="Soft Threshold (power)",ylab="Scale Free Topology Model Fit,signed R^2",type="n",
main = paste("Scale independence"));
text(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
labels=powers,cex=cex1,col="red");
# this line corresponds to using an R^2 cut-off of h
abline(h=0.90,col="red")
# Mean connectivity as a function of the soft-thresholding power
plot(sft$fitIndices[,1], sft$fitIndices[,5],
xlab="Soft Threshold (power)",ylab="Mean Connectivity", type="n",
main = paste("Mean connectivity"))
text(sft$fitIndices[,1], sft$fitIndices[,5], labels=powers, cex=cex1,col="red")
dev.off()
}
}
## step3 构建加权共表达网络(Weight co-expression network)
## 首先是一步法完成网络构建
#一步法得到net$dendrograms[[1]]
if(T){
net = blockwiseModules(
datExpr,
power = sft$powerEstimate,
maxBlockSize = 6000,
TOMType = "unsigned", minModuleSize = 30,
reassignThreshold = 0, mergeCutHeight = 0.25,
numericLabels = TRUE, pamRespectsDendro = FALSE,
saveTOMs = F,
verbose = 3
)
table(net$colors)
}
## 然后是分布法完成网络构建,仅供有探索精神的同学挑战。
## 构建加权共表达网络分为两步:
## 1. 计算邻近值,也是就是两个基因在不样品中表达量的表达相关系数(pearson correlation rho),
## 参考 2.b.2 in https://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/FemaleLiver-02-networkConstr-man.pdf
## 2. 计算topology overlap similarity (TOM)。 WGCNA认为,只通过计算两个基因的表达相关系数构建共表达网络是不足够的。
## 于是他们用TOM表示两个基因在网络结构上的相似性,即两个基因如果具有相似的邻近基因,这两个基因更倾向于有相互作用。
## 参考 2.b.3 in https://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/FemaleLiver-02-networkConstr-man.pdf
#分两步构建网络 动态修剪 融合
if(F){
#(1)网络构建 Co-expression similarity and adjacency
adjacency = adjacency(datExpr, power = sft$powerEstimate)
#(2) 邻近矩阵到拓扑矩阵的转换,Turn adjacency into topological overlap
TOM = TOMsimilarity(adjacency);
dissTOM = 1-TOM
# (3) 聚类拓扑矩阵 Call the hierarchical clustering function
geneTree = hclust(as.dist(dissTOM), method = "average");
# Plot the resulting clustering tree (dendrogram)
sizeGrWindow(12,9)
## 这个时候的geneTree与一步法的 net$dendrograms[[1]] 性质类似,但是还需要进行进一步处理
plot(geneTree, xlab="", sub="", main = "Gene clustering on TOM-based dissimilarity",
labels = FALSE, hang = 0.04);
#(4) 聚类分支的修整 dynamicTreeCut
# We like large modules, so we set the minimum module size relatively high:
minModuleSize = 30;
# Module identification using dynamic tree cut:动态修剪
dynamicMods = cutreeDynamic(dendro = geneTree, distM = dissTOM,
deepSplit = 2, pamRespectsDendro = FALSE,
minClusterSize = minModuleSize);
table(dynamicMods)
#4. 绘画结果展示
# Convert numeric lables into colors
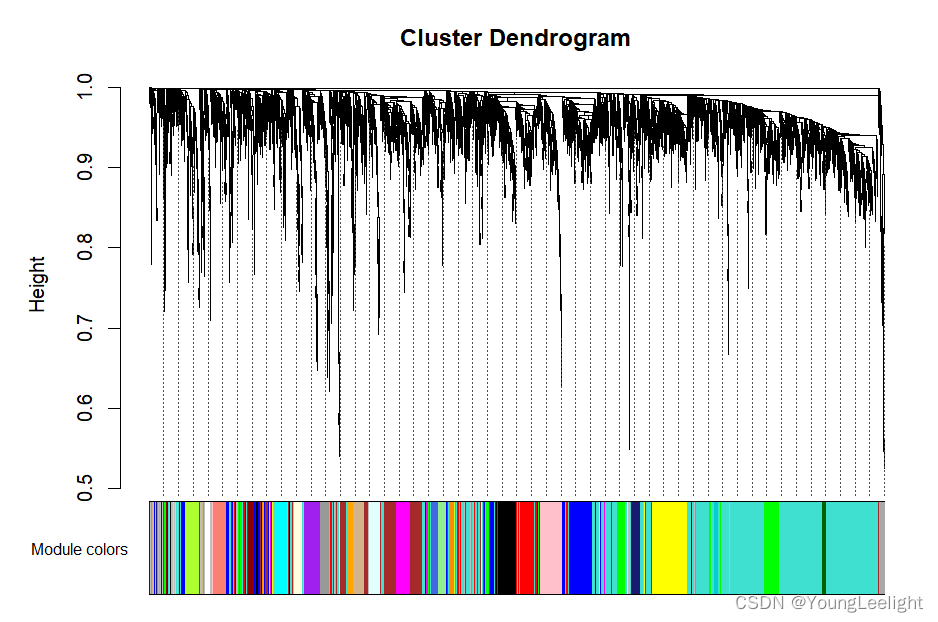
dynamicColors = labels2colors(dynamicMods)
table(dynamicColors)
# Plot the dendrogram and colors underneath
#sizeGrWindow(8,6)
plotDendroAndColors(geneTree, dynamicColors, "Dynamic Tree Cut",
dendroLabels = FALSE, hang = 0.03,
addGuide = TRUE, guideHang = 0.05,
main = "Gene dendrogram and module colors")
#5. 聚类结果相似模块的融合,Merging of modules whose expression profiles are very similar
#在聚类树中每一leaf是一个短线,代表一个基因,
#不同分之间靠的越近表示有高的共表达基因,将共表达极其相似的modules进行融合
# Calculate eigengenes
MEList = moduleEigengenes(datExpr, colors = dynamicColors)
MEs = MEList$eigengenes
# Calculate dissimilarity of module eigengenes
MEDiss = 1-cor(MEs);
# Cluster module eigengenes
METree = hclust(as.dist(MEDiss), method = "average");
# Plot the result
#sizeGrWindow(7, 6)
plot(METree, main = "Clustering of module eigengenes",
xlab = "", sub = "")
#选择有75%相关性的进行融合
MEDissThres = 0.25
# Plot the cut line into the dendrogram
abline(h=MEDissThres, col = "red")
# Call an automatic merging function
merge = mergeCloseModules(datExpr, dynamicColors, cutHeight = MEDissThres, verbose = 3)
# The merged module colors
mergedColors = merge$colors;
# Eigengenes of the new merged modules:
mergedMEs = merge$newMEs
}
## step 4 : 模块可视化
if(T){
# Convert labels to colors for plotting
mergedColors = labels2colors(net$colors)
table(mergedColors)
moduleColors=mergedColors
# Plot the dendrogram and the module colors underneath
png("step4-genes-modules.png",width = 800,height = 600)
plotDendroAndColors(net$dendrograms[[1]], mergedColors[net$blockGenes[[1]]],
"Module colors",
dendroLabels = FALSE, hang = 0.03,
addGuide = TRUE, guideHang = 0.05)
dev.off()
## assign all of the gene to their corresponding module
## hclust for the genes.
}
if(F){
#明确样本数和基因
nGenes = ncol(datExpr)
nSamples = nrow(datExpr)
#首先针对样本做个系统聚类
datExpr_tree<-hclust(dist(datExpr), method = "average")
par(mar = c(0,5,2,0))
plot(datExpr_tree, main = "Sample clustering", sub="", xlab="", cex.lab = 2,
cex.axis = 1, cex.main = 1,cex.lab=1)
## 如果这个时候样本是有性状,或者临床表型的,可以加进去看看是否聚类合理
#针对前面构造的样品矩阵添加对应颜色 ——自己要记住—— 每种颜色对那个一种样本表型,这里subtype有四个水平
sample_colors <- numbers2colors(as.numeric(factor(datTraits$subtype)), #这里非肿瘤和unknown都是green
colors = c("white","blue","red","green"),signed = FALSE)
## 这个给样品添加对应颜色的代码需要自行修改以适应自己的数据分析项目
# sample_colors <- numbers2colors( datTraits ,signed = FALSE)
## 如果样品有多种分类情况,而且 datTraits 里面都是分类信息,那么可以直接用上面代码,当然,这样给的颜色不明显,意义不大
#10个样品的系统聚类树及性状热图
par(mar = c(1,4,3,1),cex=0.8)
png("sample-subtype-cluster.png",width = 800,height = 600)
plotDendroAndColors(datExpr_tree, sample_colors,
groupLabels = colnames(sample),
cex.dendroLabels = 0.8,
marAll = c(1, 4, 3, 1),
cex.rowText = 0.01,
main = "Sample dendrogram and trait heatmap")
dev.off()
}
## step 5 (最重要的) 模块和性状的关系
## 这一步主要是针对于连续变量,如果是分类变量,需要转换成连续变量方可使用
table(datTraits$subtype)
if(T){
nGenes = ncol(datExpr)
nSamples = nrow(datExpr)
design=model.matrix(~0+ datTraits$subtype)
colnames(design)=levels(datTraits$subtype)
moduleColors <- labels2colors(net$colors)
# Recalculate MEs with color labels
MEs0 = moduleEigengenes(datExpr, moduleColors)$eigengenes
MEs = orderMEs(MEs0); ##不同颜色的模块的ME值矩 (样本vs模块)
moduleTraitCor = cor(MEs, design , use = "p");
moduleTraitPvalue = corPvalueStudent(moduleTraitCor, nSamples)
sizeGrWindow(10,6)
# Will display correlations and their p-values
textMatrix = paste(signif(moduleTraitCor, 2), "\n(",
signif(moduleTraitPvalue, 1), ")", sep = "");
dim(textMatrix) = dim(moduleTraitCor)
png("step5-Module-trait-relationships.png",width = 800,height = 1200,res = 120)
par(mar = c(6, 8.5, 3, 3));
# Display the correlation values within a heatmap plot
labeledHeatmap(Matrix = moduleTraitCor,
xLabels = colnames(design),
yLabels = names(MEs),
ySymbols = names(MEs),
colorLabels = FALSE,
colors = greenWhiteRed(50),
textMatrix = textMatrix,
setStdMargins = FALSE,
cex.text = 0.5,
zlim = c(-1,1),
main = paste("Module-trait relationships"))
dev.off()
# 除了上面的热图展现形状与基因模块的相关性外
# 还可以是条形图,但是只能是指定某个形状
# 或者自己循环一下批量出图。
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
y=Luminal
GS1=as.numeric(cor(y,datExpr, use="p"))
GeneSignificance=abs(GS1)
# Next module significance is defined as average gene significance.
ModuleSignificance=tapply(GeneSignificance,
moduleColors, mean, na.rm=T)
sizeGrWindow(8,7)
par(mfrow = c(1,1))
# 如果模块太多,下面的展示就不友好
# 不过,我们可以自定义出图。
plotModuleSignificance(GeneSignificance,moduleColors)
}
## step 6 (第二重要的):感兴趣性状的模块的具体基因分析
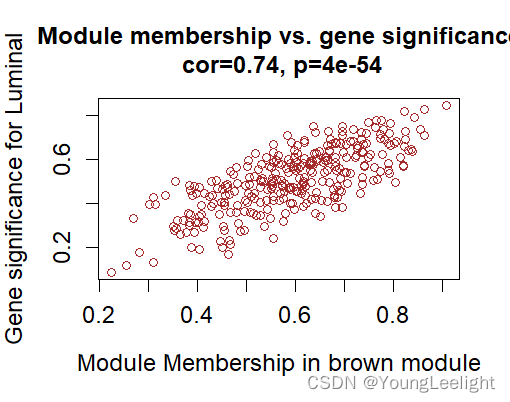
# 查看第五步出图:step5-Module-trait-relationships.png
# 发现跟 Luminal 亚型 最相关的是 brown 模块
# 所以接下来就分析这两个
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
module = "brown"
if(T){
# names (colors) of the modules
modNames = substring(names(MEs), 3)
geneModuleMembership = as.data.frame(cor(datExpr, MEs, use = "p"));
## 算出每个模块跟基因的皮尔森相关系数矩
## MEs是每个模块在每个样本里面的
## datExpr是每个基因在每个样本的表达量
MMPvalue = as.data.frame(corPvalueStudent(as.matrix(geneModuleMembership), nSamples));
names(geneModuleMembership) = paste("MM", modNames, sep="");
names(MMPvalue) = paste("p.MM", modNames, sep="");
geneModuleMembership[1:4,1:4]
## 只有连续型性状才能只有计算
## 这里把是否属 Luminal 表型这个变量0,1进行数值化
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
geneTraitSignificance = as.data.frame(cor(datExpr, Luminal, use = "p"));
GSPvalue = as.data.frame(corPvalueStudent(as.matrix(geneTraitSignificance), nSamples));
names(geneTraitSignificance) = paste("GS.", names(Luminal), sep="");
names(GSPvalue) = paste("p.GS.", names(Luminal), sep="");
module = "brown"
column = match(module, modNames);
moduleGenes = moduleColors==module;
png("step6-Module_membership-gene_significance.png",width = 800,height = 600)
#sizeGrWindow(7, 7);
par(mfrow = c(1,1));
verboseScatterplot(abs(geneModuleMembership[moduleGenes, column]),
abs(geneTraitSignificance[moduleGenes, 1]),
xlab = paste("Module Membership in", module, "module"),
ylab = "Gene significance for Luminal",
main = paste("Module membership vs. gene significance\n"),
cex.main = 1.2, cex.lab = 1.2, cex.axis = 1.2, col = module)
dev.off()
}
## step 7
# 主要是可视化 TOM矩阵,WGCNA的标准配图
# 然后可视化不同 模块 的相关性 热图
# 不同模块的层次聚类图
# 还有模块诊断,主要是 intramodular connectivity
if(T){
nGenes = ncol(datExpr)
nSamples = nrow(datExpr)
geneTree = net$dendrograms[[1]];
dissTOM = 1-TOMsimilarityFromExpr(datExpr, power = 6);
plotTOM = dissTOM^7;
diag(plotTOM) = NA;
#TOMplot(plotTOM, geneTree, moduleColors, main = "Network heatmap plot, all genes")
nSelect = 400
# For reproducibility, we set the random seed
set.seed(10);
select = sample(nGenes, size = nSelect);
selectTOM = dissTOM[select, select];
# There’s no simple way of restricting a clustering tree to a subset of genes, so we must re-cluster.
selectTree = hclust(as.dist(selectTOM), method = "average")
selectColors = moduleColors[select];
# Open a graphical window
sizeGrWindow(9,9)
# Taking the dissimilarity to a power, say 10, makes the plot more informative by effectively changing
# the color palette; setting the diagonal to NA also improves the clarity of the plot
plotDiss = selectTOM^7;
diag(plotDiss) = NA;
png("step7-Network-heatmap.png",width = 800,height = 600)
TOMplot(plotDiss, selectTree, selectColors, main = "Network heatmap plot, selected genes")
dev.off()
# Recalculate module eigengenes
MEs = moduleEigengenes(datExpr, moduleColors)$eigengenes
## 只有连续型性状才能只有计算
## 这里把是否属 Luminal 表型这个变量0,1进行数值化
Luminal = as.data.frame(design[,3]);
names(Luminal) = "Luminal"
# Add the weight to existing module eigengenes
MET = orderMEs(cbind(MEs, Luminal))
# Plot the relationships among the eigengenes and the trait
sizeGrWindow(5,7.5);
par(cex = 0.9)
png("step7-Eigengene-dendrogram.png",width = 800,height = 600)
plotEigengeneNetworks(MET, "", marDendro = c(0,4,1,2), marHeatmap = c(3,4,1,2), cex.lab = 0.8, xLabelsAngle
= 90)
dev.off()
# Plot the dendrogram
sizeGrWindow(6,6);
par(cex = 1.0)
## 模块的进化树
png("step7-Eigengene-dendrogram-hclust.png",width = 800,height = 600)
plotEigengeneNetworks(MET, "Eigengene dendrogram", marDendro = c(0,4,2,0),
plotHeatmaps = FALSE)
dev.off()
# Plot the heatmap matrix (note: this plot will overwrite the dendrogram plot)
par(cex = 1.0)
## 性状与模块热
png("step7-Eigengene-adjacency-heatmap.png",width = 800,height = 600)
plotEigengeneNetworks(MET, "Eigengene adjacency heatmap", marHeatmap = c(3,4,2,2),
plotDendrograms = FALSE, xLabelsAngle = 90)
dev.off()
}
## step 8
# 主要是关心具体某个模块内部的基因
if(T){
# Select module
module = "brown";
# Select module probes
probes = colnames(datExpr) ## 我们例子里面的probe就是基因
inModule = (moduleColors==module);
modProbes = probes[inModule];
head(modProbes)
# 如果使用WGCNA包自带的热图就很丑。
which.module="brown";
dat=datExpr[,moduleColors==which.module ]
plotMat(t(scale(dat)),nrgcols=30,rlabels=T,
clabels=T,rcols=which.module,
title=which.module )
datExpr[1:4,1:4]
dat=t(datExpr[,moduleColors==which.module ] )
library(pheatmap)
pheatmap(dat ,show_colnames =F,show_rownames = F) #对那些提取出来的1000个基因所在的每一行取出,组合起来为一个新的表达矩阵
n=t(scale(t(log(dat+1)))) # 'scale'可以对log-ratio数值进行归一化
n[n>2]=2
n[n< -2]= -2
n[1:4,1:4]
pheatmap(n,show_colnames =F,show_rownames = F)
group_list=datTraits$subtype
ac=data.frame(g=group_list)
rownames(ac)=colnames(n)
pheatmap(n,show_colnames =F,show_rownames = F,
annotation_col=ac )
# 可以很清晰的看到,所有的形状相关的模块基因
# 其实未必就不是差异表达基因。
}
## step 9
# 导出模块内部基因的连接关系,进入其它可视化软件
# 比如 cytoscape软件等等。
if(T){
# Recalculate topological overlap
TOM = TOMsimilarityFromExpr(datExpr, power = 6);
# Select module
module = "brown";
# Select module probes
probes = colnames(datExpr) ## 我们例子里面的probe就是基因
inModule = (moduleColors==module);
modProbes = probes[inModule];
## 也是提取指定模块的基因名
# Select the corresponding Topological Overlap
modTOM = TOM[inModule, inModule];
dimnames(modTOM) = list(modProbes, modProbes)
## 模块对应的基因关系矩
cyt = exportNetworkToCytoscape(
modTOM,
edgeFile = paste("CytoscapeInput-edges-", paste(module, collapse="-"), ".txt", sep=""),
nodeFile = paste("CytoscapeInput-nodes-", paste(module, collapse="-"), ".txt", sep=""),
weighted = TRUE,
threshold = 0.02,
nodeNames = modProbes,
nodeAttr = moduleColors[inModule]
);
}