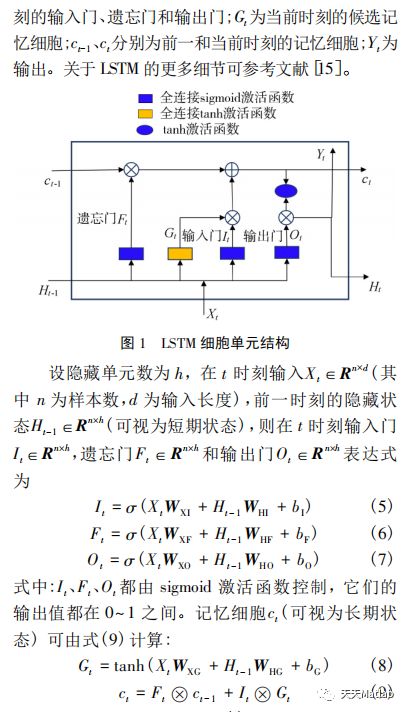
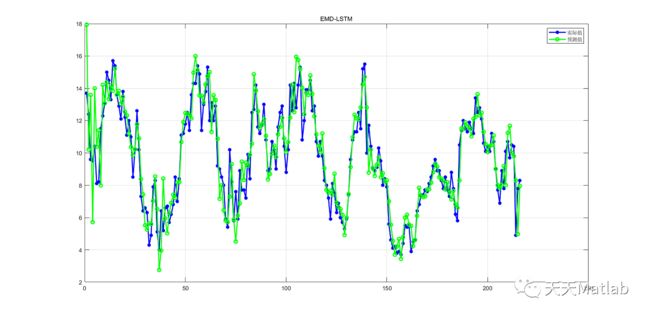
【LSTM时序预测】基于EMD结合长短时记忆网络LSTM实现风速数据预测Matlab源码
✅作者简介:热爱科研的Matlab仿真开发者,修心和技术同步精进,matlab项目合作可私信。
个人主页:Matlab科研工作室
个人信条:格物致知。
更多Matlab仿真内容点击
智能优化算法 神经网络预测 雷达通信 无线传感器
信号处理 图像处理 路径规划 元胞自动机 无人机 电力系统
⛄ 内容介绍
⛄ 部分代码
%EMD computes Empirical Mode Decomposition
%这个是通过设定阈值来确定分解出的imf个数
%
% Syntax
%
%
% IMF = EMD(X)
% IMF = EMD(X,...,'Option_name',Option_value,...)
% IMF = EMD(X,OPTS)
% [IMF,ORT,NB_ITERATIONS] = EMD(...)
%
%
% Description
%
%
% IMF = EMD(X) where X is a real vector computes the Empirical Mode
% Decomposition [1] of X, resulting in a matrix IMF containing 1 IMF per row, the
% last one being the residue. The default stopping criterion is the one proposed
% in [2]:
%
% at each point, mean_amplitude < THRESHOLD2*envelope_amplitude
% &
% mean of boolean array {(mean_amplitude)/(envelope_amplitude) > THRESHOLD} < TOLERANCE
% &
% |#zeros-#extrema|<=1
%
% where mean_amplitude = abs(envelope_max+envelope_min)/2
% and envelope_amplitude = abs(envelope_max-envelope_min)/2
%
% IMF = EMD(X) where X is a complex vector computes Bivariate Empirical Mode
% Decomposition [3] of X, resulting in a matrix IMF containing 1 IMF per row, the
% last one being the residue. The default stopping criterion is similar to the
% one proposed in [2]:
%
% at each point, mean_amplitude < THRESHOLD2*envelope_amplitude
% &
% mean of boolean array {(mean_amplitude)/(envelope_amplitude) > THRESHOLD} < TOLERANCE
%
% where mean_amplitude and envelope_amplitude have definitions similar to the
% real case
%
% IMF = EMD(X,...,'Option_name',Option_value,...) sets options Option_name to
% the specified Option_value (see Options)
%
% IMF = EMD(X,OPTS) is equivalent to the above syntax provided OPTS is a struct
% object with field names corresponding to option names and field values being the
% associated values
%
% [IMF,ORT,NB_ITERATIONS] = EMD(...) returns an index of orthogonality
% ________
% _ |IMF(i,:).*IMF(j,:)|
% ORT = \ _____________________
% /
% ? || X ||?% i~=j
%
% and the number of iterations to extract each mode in NB_ITERATIONS
%
%
% Options
%
%
% stopping criterion options:
%
% STOP: vector of stopping parameters [THRESHOLD,THRESHOLD2,TOLERANCE]
% if the input vector's length is less than 3, only the first parameters are
% set, the remaining ones taking default values.
% default: [0.05,0.5,0.05]
%
% FIX (int): disable the default stopping criterion and do exactly
% number of sifting iterations for each mode
%
% FIX_H (int): disable the default stopping criterion and do
% iterations with |#zeros-#extrema|<=1 to stop [4]
%
% bivariate/complex EMD options:
%
% COMPLEX_VERSION: selects the algorithm used for complex EMD ([3])
% COMPLEX_VERSION = 1: "algorithm 1"
% COMPLEX_VERSION = 2: "algorithm 2" (default)
%
% NDIRS: number of directions in which envelopes are computed (default 4)
% rem: the actual number of directions (according to [3]) is 2*NDIRS
%
% other options:
%
% T: sampling times (line vector) (default: 1:length(x))
%
% MAXITERATIONS: maximum number of sifting iterations for the computation of each
% mode (default: 2000)
%
% MAXMODES: maximum number of imfs extracted (default: Inf)
%
% DISPLAY: if equals to 1 shows sifting steps with pause
% if equals to 2 shows sifting steps without pause (movie style)
% rem: display is disabled when the input is complex
%
% INTERP: interpolation scheme: 'linear', 'cubic', 'pchip' or 'spline' (default)
% see interp1 documentation for details
%
% MASK: masking signal used to improve the decomposition according to [5]
%
%
% Examples
%
%
%X = rand(1,512);
%
%IMF = emd(X);
%
%IMF = emd(X,'STOP',[0.1,0.5,0.05],'MAXITERATIONS',100);
%
%T=linspace(0,20,1e3);
%X = 2*exp(i*T)+exp(3*i*T)+.5*T;
%IMF = emd(X,'T',T);
%
%OPTIONS.DISLPAY = 1;
%OPTIONS.FIX = 10;
%OPTIONS.MAXMODES = 3;
%[IMF,ORT,NBITS] = emd(X,OPTIONS);
%
%
% References
%
%
% [1] N. E. Huang et al., "The empirical mode decomposition and the
% Hilbert spectrum for non-linear and non stationary time series analysis",
% Proc. Royal Soc. London A, Vol. 454, pp. 903-995, 1998
%
% [2] G. Rilling, P. Flandrin and P. Gon鏰lves
% "On Empirical Mode Decomposition and its algorithms",
% IEEE-EURASIP Workshop on Nonlinear Signal and Image Processing
% NSIP-03, Grado (I), June 2003
%
% [3] G. Rilling, P. Flandrin, P. Gon鏰lves and J. M. Lilly.,
% "Bivariate Empirical Mode Decomposition",
% Signal Processing Letters (submitted)
%
% [4] N. E. Huang et al., "A confidence limit for the Empirical Mode
% Decomposition and Hilbert spectral analysis",
% Proc. Royal Soc. London A, Vol. 459, pp. 2317-2345, 2003
%
% [5] R. Deering and J. F. Kaiser, "The use of a masking signal to improve
% empirical mode decomposition", ICASSP 2005
%
%
% See also
% emd_visu (visualization),
% emdc, emdc_fix (fast implementations of EMD),
% cemdc, cemdc_fix, cemdc2, cemdc2_fix (fast implementations of bivariate EMD),
% hhspectrum (Hilbert-Huang spectrum)
%
%
% G. Rilling, last modification: 3.2007
function [imf,ort,nbits] = emd(varargin)
[x,t,sd,sd2,tol,MODE_COMPLEX,ndirs,display_sifting,sdt,sd2t,r,imf,k,nbit,NbIt,MAXITERATIONS,FIXE,FIXE_H,MAXMODES,INTERP,mask] = init(varargin{:});
if display_sifting
fig_h = figure;
end
%main loop : requires at least 3 extrema to proceed
while (~stop_EMD(r,MODE_COMPLEX,ndirs) && (k < MAXMODES+1 || MAXMODES == 0) && ~any(mask))
% current mode
m = r;
% mode at previous iteration
mp = m;
%computation of mean and stopping criterion
if FIXE
[stop_sift,moyenne] = stop_sifting_fixe(t,m,INTERP,MODE_COMPLEX,ndirs);
elseif FIXE_H
stop_count = 0;
[stop_sift,moyenne] = stop_sifting_fixe_h(t,m,INTERP,stop_count,FIXE_H,MODE_COMPLEX,ndirs);
else
[stop_sift,moyenne] = stop_sifting(m,t,sd,sd2,tol,INTERP,MODE_COMPLEX,ndirs);
end
% in case the current mode is so small that machine precision can cause
% spurious extrema to appear
if (max(abs(m))) < (1e-10)*(max(abs(x)))
if ~stop_sift
warning('emd:warning','forced stop of EMD : too small amplitude')
else
disp('forced stop of EMD : too small amplitude')
end
break
end
% sifting loop
while ~stop_sift && nbit if(~MODE_COMPLEX && nbit>MAXITERATIONS/5 && mod(nbit,floor(MAXITERATIONS/10))==0 && ~FIXE && nbit > 100) disp(['mode ',int2str(k),', iteration ',int2str(nbit)]) if exist('s','var') disp(['stop parameter mean value : ',num2str(s)]) end [im,iM] = extr(m); disp([int2str(sum(m(im) > 0)),' minima > 0; ',int2str(sum(m(iM) < 0)),' maxima < 0.']) end %sifting m = m - moyenne; %computation of mean and stopping criterion if FIXE [stop_sift,moyenne] = stop_sifting_fixe(t,m,INTERP,MODE_COMPLEX,ndirs); elseif FIXE_H [stop_sift,moyenne,stop_count] = stop_sifting_fixe_h(t,m,INTERP,stop_count,FIXE_H,MODE_COMPLEX,ndirs); else [stop_sift,moyenne,s] = stop_sifting(m,t,sd,sd2,tol,INTERP,MODE_COMPLEX,ndirs); end % display if display_sifting && ~MODE_COMPLEX NBSYM = 2; [indmin,indmax] = extr(mp); [tmin,tmax,mmin,mmax] = boundary_conditions(indmin,indmax,t,mp,mp,NBSYM); envminp = interp1(tmin,mmin,t,INTERP); envmaxp = interp1(tmax,mmax,t,INTERP); envmoyp = (envminp+envmaxp)/2; if FIXE || FIXE_H display_emd_fixe(t,m,mp,r,envminp,envmaxp,envmoyp,nbit,k,display_sifting) else sxp=2*(abs(envmoyp))./(abs(envmaxp-envminp)); sp = mean(sxp); display_emd(t,m,mp,r,envminp,envmaxp,envmoyp,s,sp,sxp,sdt,sd2t,nbit,k,display_sifting,stop_sift) end end mp = m; nbit=nbit+1; NbIt=NbIt+1; if(nbit==(MAXITERATIONS-1) && ~FIXE && nbit > 100) if exist('s','var') warning('emd:warning',['forced stop of sifting : too many iterations... mode ',int2str(k),'. stop parameter mean value : ',num2str(s)]) else warning('emd:warning',['forced stop of sifting : too many iterations... mode ',int2str(k),'.']) end end end % sifting loop imf(k,:) = m; if display_sifting disp(['mode ',int2str(k),' stored']) end nbits(k) = nbit; k = k+1; r = r - m; nbit=0; end %main loop if any(r) && ~any(mask) imf(k,:) = r; end ort = io(x,imf); if display_sifting close end end %--------------------------------------------------------------------------------------------------- % tests if there are enough (3) extrema to continue the decomposition function stop = stop_EMD(r,MODE_COMPLEX,ndirs) if MODE_COMPLEX for k = 1:ndirs phi = (k-1)*pi/ndirs; [indmin,indmax] = extr(real(exp(i*phi)*r)); ner(k) = length(indmin) + length(indmax); end stop = any(ner < 3); else [indmin,indmax] = extr(r); ner = length(indmin) + length(indmax); stop = ner < 3; end end %--------------------------------------------------------------------------------------------------- % computes the mean of the envelopes and the mode amplitude estimate function [envmoy,nem,nzm,amp] = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs) NBSYM = 2; if MODE_COMPLEX switch MODE_COMPLEX case 1 for k = 1:ndirs phi = (k-1)*pi/ndirs; y = real(exp(-i*phi)*m); [indmin,indmax,indzer] = extr(y); nem(k) = length(indmin)+length(indmax); nzm(k) = length(indzer); [tmin,tmax,zmin,zmax] = boundary_conditions(indmin,indmax,t,y,m,NBSYM); envmin(k,:) = interp1(tmin,zmin,t,INTERP); envmax(k,:) = interp1(tmax,zmax,t,INTERP); end envmoy = mean((envmin+envmax)/2,1); if nargout > 3 amp = mean(abs(envmax-envmin),1)/2; end case 2 for k = 1:ndirs phi = (k-1)*pi/ndirs; y = real(exp(-i*phi)*m); [indmin,indmax,indzer] = extr(y); nem(k) = length(indmin)+length(indmax); nzm(k) = length(indzer); [tmin,tmax,zmin,zmax] = boundary_conditions(indmin,indmax,t,y,y,NBSYM); envmin(k,:) = exp(i*phi)*interp1(tmin,zmin,t,INTERP); envmax(k,:) = exp(i*phi)*interp1(tmax,zmax,t,INTERP); end envmoy = mean((envmin+envmax),1); if nargout > 3 amp = mean(abs(envmax-envmin),1)/2; end end else [indmin,indmax,indzer] = extr(m); nem = length(indmin)+length(indmax); nzm = length(indzer); [tmin,tmax,mmin,mmax] = boundary_conditions(indmin,indmax,t,m,m,NBSYM); envmin = interp1(tmin,mmin,t,INTERP); envmax = interp1(tmax,mmax,t,INTERP); envmoy = (envmin+envmax)/2; if nargout > 3 amp = mean(abs(envmax-envmin),1)/2; end end end %------------------------------------------------------------------------------- % default stopping criterion function [stop,envmoy,s] = stop_sifting(m,t,sd,sd2,tol,INTERP,MODE_COMPLEX,ndirs) try [envmoy,nem,nzm,amp] = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs); sx = abs(envmoy)./amp; s = mean(sx); stop = ~((mean(sx > sd) > tol | any(sx > sd2)) & (all(nem > 2))); if ~MODE_COMPLEX stop = stop && ~(abs(nzm-nem)>1); end catch stop = 1; envmoy = zeros(1,length(m)); s = NaN; end end %------------------------------------------------------------------------------- % stopping criterion corresponding to option FIX function [stop,moyenne]= stop_sifting_fixe(t,m,INTERP,MODE_COMPLEX,ndirs) try moyenne = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs); stop = 0; catch moyenne = zeros(1,length(m)); stop = 1; end end %------------------------------------------------------------------------------- % stopping criterion corresponding to option FIX_H function [stop,moyenne,stop_count]= stop_sifting_fixe_h(t,m,INTERP,stop_count,FIXE_H,MODE_COMPLEX,ndirs) try [moyenne,nem,nzm] = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs); if (all(abs(nzm-nem)>1)) stop = 0; stop_count = 0; else stop_count = stop_count+1; stop = (stop_count == FIXE_H); end catch moyenne = zeros(1,length(m)); stop = 1; end end %------------------------------------------------------------------------------- % displays the progression of the decomposition with the default stopping criterion function display_emd(t,m,mp,r,envmin,envmax,envmoy,s,sb,sx,sdt,sd2t,nbit,k,display_sifting,stop_sift) subplot(4,1,1) plot(t,mp);hold on; plot(t,envmax,'--k');plot(t,envmin,'--k');plot(t,envmoy,'r'); title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' before sifting']); set(gca,'XTick',[]) hold off subplot(4,1,2) plot(t,sx) hold on plot(t,sdt,'--r') plot(t,sd2t,':k') title('stop parameter') set(gca,'XTick',[]) hold off subplot(4,1,3) plot(t,m) title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' after sifting']); set(gca,'XTick',[]) subplot(4,1,4); plot(t,r-m) title('residue'); disp(['stop parameter mean value : ',num2str(sb),' before sifting and ',num2str(s),' after']) if stop_sift disp('last iteration for this mode') end if display_sifting == 2 pause(0.01) else pause end end %--------------------------------------------------------------------------------------------------- % displays the progression of the decomposition with the FIX and FIX_H stopping criteria function display_emd_fixe(t,m,mp,r,envmin,envmax,envmoy,nbit,k,display_sifting) subplot(3,1,1) plot(t,mp);hold on; plot(t,envmax,'--k');plot(t,envmin,'--k');plot(t,envmoy,'r'); title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' before sifting']); set(gca,'XTick',[]) hold off subplot(3,1,2) plot(t,m) title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' after sifting']); set(gca,'XTick',[]) subplot(3,1,3); plot(t,r-m) title('residue'); if display_sifting == 2 pause(0.01) else pause end end %--------------------------------------------------------------------------------------- % defines new extrema points to extend the interpolations at the edges of the % signal (mainly mirror symmetry) function [tmin,tmax,zmin,zmax] = boundary_conditions(indmin,indmax,t,x,z,nbsym) lx = length(x); if (length(indmin) + length(indmax) < 3) error('not enough extrema') end % boundary conditions for interpolations : if indmax(1) < indmin(1) if x(1) > x(indmin(1)) lmax = fliplr(indmax(2:min(end,nbsym+1))); lmin = fliplr(indmin(1:min(end,nbsym))); lsym = indmax(1); else lmax = fliplr(indmax(1:min(end,nbsym))); lmin = [fliplr(indmin(1:min(end,nbsym-1))),1]; lsym = 1; end else if x(1) < x(indmax(1)) lmax = fliplr(indmax(1:min(end,nbsym))); lmin = fliplr(indmin(2:min(end,nbsym+1))); lsym = indmin(1); else lmax = [fliplr(indmax(1:min(end,nbsym-1))),1]; lmin = fliplr(indmin(1:min(end,nbsym))); lsym = 1; end end if indmax(end) < indmin(end) if x(end) < x(indmax(end)) rmax = fliplr(indmax(max(end-nbsym+1,1):end)); rmin = fliplr(indmin(max(end-nbsym,1):end-1)); rsym = indmin(end); else rmax = [lx,fliplr(indmax(max(end-nbsym+2,1):end))]; rmin = fliplr(indmin(max(end-nbsym+1,1):end)); rsym = lx; end else if x(end) > x(indmin(end)) rmax = fliplr(indmax(max(end-nbsym,1):end-1)); rmin = fliplr(indmin(max(end-nbsym+1,1):end)); rsym = indmax(end); else rmax = fliplr(indmax(max(end-nbsym+1,1):end)); rmin = [lx,fliplr(indmin(max(end-nbsym+2,1):end))]; rsym = lx; end end tlmin = 2*t(lsym)-t(lmin); tlmax = 2*t(lsym)-t(lmax); trmin = 2*t(rsym)-t(rmin); trmax = 2*t(rsym)-t(rmax); % in case symmetrized parts do not extend enough if tlmin(1) > t(1) || tlmax(1) > t(1) if lsym == indmax(1) lmax = fliplr(indmax(1:min(end,nbsym))); else lmin = fliplr(indmin(1:min(end,nbsym))); end if lsym == 1 error('bug') end lsym = 1; tlmin = 2*t(lsym)-t(lmin); tlmax = 2*t(lsym)-t(lmax); end if trmin(end) < t(lx) || trmax(end) < t(lx) if rsym == indmax(end) rmax = fliplr(indmax(max(end-nbsym+1,1):end)); else rmin = fliplr(indmin(max(end-nbsym+1,1):end)); end if rsym == lx error('bug') end rsym = lx; trmin = 2*t(rsym)-t(rmin); trmax = 2*t(rsym)-t(rmax); end zlmax =z(lmax); zlmin =z(lmin); zrmax =z(rmax); zrmin =z(rmin); tmin = [tlmin t(indmin) trmin]; tmax = [tlmax t(indmax) trmax]; zmin = [zlmin z(indmin) zrmin]; zmax = [zlmax z(indmax) zrmax]; end %--------------------------------------------------------------------------------------------------- %extracts the indices of extrema function [indmin, indmax, indzer] = extr(x,t) if(nargin==1) t=1:length(x); end m = length(x); if nargout > 2 x1=x(1:m-1); x2=x(2:m); indzer = find(x1.*x2<0); if any(x == 0) iz = find( x==0 ); indz = []; if any(diff(iz)==1) zer = x == 0; dz = diff([0 zer 0]); debz = find(dz == 1); finz = find(dz == -1)-1; indz = round((debz+finz)/2); else indz = iz; end indzer = sort([indzer indz]); end end d = diff(x); n = length(d); d1 = d(1:n-1); d2 = d(2:n); indmin = find(d1.*d2<0 & d1<0)+1; indmax = find(d1.*d2<0 & d1>0)+1; % when two or more successive points have the same value we consider only one extremum in the middle of the constant area % (only works if the signal is uniformly sampled) if any(d==0) imax = []; imin = []; bad = (d==0); dd = diff([0 bad 0]); debs = find(dd == 1); fins = find(dd == -1); if debs(1) == 1 if length(debs) > 1 debs = debs(2:end); fins = fins(2:end); else debs = []; fins = []; end end if length(debs) > 0 if fins(end) == m if length(debs) > 1 debs = debs(1:(end-1)); fins = fins(1:(end-1)); else debs = []; fins = []; end end end lc = length(debs); if lc > 0 for k = 1:lc if d(debs(k)-1) > 0 if d(fins(k)) < 0 imax = [imax round((fins(k)+debs(k))/2)]; end else if d(fins(k)) > 0 imin = [imin round((fins(k)+debs(k))/2)]; end end end end if length(imax) > 0 indmax = sort([indmax imax]); end if length(imin) > 0 indmin = sort([indmin imin]); end end end %--------------------------------------------------------------------------------------------------- function ort = io(x,imf) % ort = IO(x,imf) computes the index of orthogonality % % inputs : - x : analyzed signal % - imf : empirical mode decomposition n = size(imf,1); s = 0; for i = 1:n for j =1:n if i~=j s = s + abs(sum(imf(i,:).*conj(imf(j,:)))/sum(x.^2)); end end end ort = 0.5*s; end %--------------------------------------------------------------------------------------------------- function [x,t,sd,sd2,tol,MODE_COMPLEX,ndirs,display_sifting,sdt,sd2t,r,imf,k,nbit,NbIt,MAXITERATIONS,FIXE,FIXE_H,MAXMODES,INTERP,mask] = init(varargin) x = varargin{1}; if nargin == 2 if isstruct(varargin{2}) inopts = varargin{2}; else error('when using 2 arguments the first one is the analyzed signal X and the second one is a struct object describing the options') end elseif nargin > 2 try inopts = struct(varargin{2:end}); catch error('bad argument syntax') end end % default for stopping defstop = [0.05,0.5,0.05]; opt_fields = {'t','stop','display','maxiterations','fix','maxmodes','interp','fix_h','mask','ndirs','complex_version'}; defopts.stop = defstop; defopts.display = 0; defopts.t = 1:max(size(x)); defopts.maxiterations = 2000; defopts.fix = 0; defopts.maxmodes = 0; defopts.interp = 'spline'; defopts.fix_h = 0; defopts.mask = 0; defopts.ndirs = 4; defopts.complex_version = 2; opts = defopts; if(nargin==1) inopts = defopts; elseif nargin == 0 error('not enough arguments') end names = fieldnames(inopts); for nom = names' if ~any(strcmpi(char(nom), opt_fields)) error(['bad option field name: ',char(nom)]) end if ~isempty(eval(['inopts.',char(nom)])) % empty values are discarded eval(['opts.',lower(char(nom)),' = inopts.',char(nom),';']) end end t = opts.t; stop = opts.stop; display_sifting = opts.display; MAXITERATIONS = opts.maxiterations; FIXE = opts.fix; MAXMODES = opts.maxmodes; INTERP = opts.interp; FIXE_H = opts.fix_h; mask = opts.mask; ndirs = opts.ndirs; complex_version = opts.complex_version; if ~isvector(x) error('X must have only one row or one column') end if size(x,1) > 1 x = x.'; end if ~isvector(t) error('option field T must have only one row or one column') end if ~isreal(t) error('time instants T must be a real vector') end if size(t,1) > 1 t = t'; end if (length(t)~=length(x)) error('X and option field T must have the same length') end if ~isvector(stop) || length(stop) > 3 error('option field STOP must have only one row or one column of max three elements') end if ~all(isfinite(x)) error('data elements must be finite') end if size(stop,1) > 1 stop = stop'; end L = length(stop); if L < 3 stop(3)=defstop(3); end if L < 2 stop(2)=defstop(2); end if ~ischar(INTERP) || ~any(strcmpi(INTERP,{'linear','cubic','spline'})) error('INTERP field must be ''linear'', ''cubic'', ''pchip'' or ''spline''') end %special procedure when a masking signal is specified if any(mask) if ~isvector(mask) || length(mask) ~= length(x) error('masking signal must have the same dimension as the analyzed signal X') end if size(mask,1) > 1 mask = mask.'; end opts.mask = 0; imf1 = emd(x+mask,opts); imf2 = emd(x-mask,opts); if size(imf1,1) ~= size(imf2,1) warning('emd:warning',['the two sets of IMFs have different sizes: ',int2str(size(imf1,1)),' and ',int2str(size(imf2,1)),' IMFs.']) end S1 = size(imf1,1); S2 = size(imf2,1); if S1 ~= S2 if S1 < S2 tmp = imf1; imf1 = imf2; imf2 = tmp; end imf2(max(S1,S2),1) = 0; end imf = (imf1+imf2)/2; end sd = stop(1); sd2 = stop(2); tol = stop(3); lx = length(x); sdt = sd*ones(1,lx); sd2t = sd2*ones(1,lx); if FIXE MAXITERATIONS = FIXE; if FIXE_H error('cannot use both ''FIX'' and ''FIX_H'' modes') end end MODE_COMPLEX = ~isreal(x)*complex_version; if MODE_COMPLEX && complex_version ~= 1 && complex_version ~= 2 error('COMPLEX_VERSION parameter must equal 1 or 2') end % number of extrema and zero-crossings in residual ner = lx; nzr = lx; r = x; if ~any(mask) % if a masking signal is specified "imf" already exists at this stage imf = []; end k = 1; % iterations counter for extraction of 1 mode nbit=0; % total iterations counter NbIt=0; end %--------------------------------------------------------------------------------------------------- [1]王亦斌、孙涛、梁雪春、谢海洋. “基于EMD-LSTM模型的河水量水位预测。” 水利水电科技进展 40.6(2020):8. ❤️部分理论引用网络文献,若有侵权联系博主删除 ❤️ 关注我领取海量matlab电子书和数学建模资料⛄ 运行结果
⛄ 参考文献
⛄ Matlab代码关注