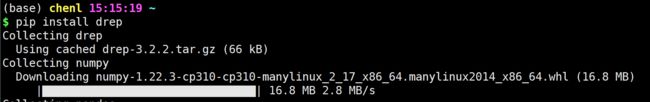
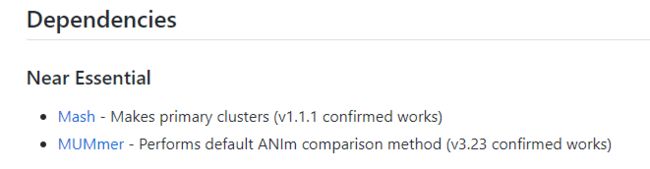
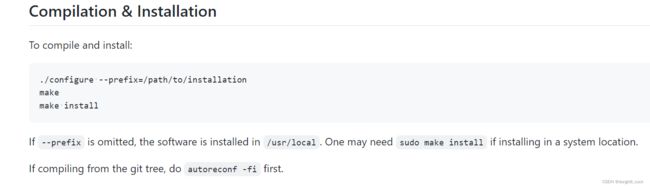
- 《CPython Internals》阅读笔记:p96-p96
python
《CPythonInternals》学习第6天,p96-p96总结,总计1页。一、技术总结1.parser-tokenizerp92,Creatingaconcretesyntaxtreeusingaparser-tokenizer,orlexer.p96,CPythonhasaparser-tokenizermodule,writteninC.当做这在92页提到parser-tokenizer的
- 深度学习每周学习总结R4(LSTM-实现糖尿病探索与预测)
大地之灯
每周深度学习总结深度学习学习lstm人工智能算法
本文为365天深度学习训练营中的学习记录博客R6中的内容,为了便于自己整理总结起名为R4原作者:K同学啊|接辅导、项目定制目录0.总结1.LSTM介绍LSTM的基本组成部分如何理解与应用LSTM2.数据预处理3.数据集构建4.定义模型5.初始化模型及优化器6.训练函数7.测试函数8.训练过程9.模型评估0.总结数据导入及处理部分:在PyTorch中,我们通常先将NumPy数组转换为torch.Te
- Python列表方法
L_lemo004
Pythonpython
目录添加元素Pythonappend()方法添加元素Pythonextend()方法添加元素Pythoninsert()方法插入元素删除元素del:根据索引值删除元素pop():根据索引值删除元素remove():根据元素值进行删除clear():删除列表所有元素修改元素修改单个元素修改一组元素查找元素index()方法count()方法添加元素实际开发中,经常需要对Python列表进行更新,包括
- requests库的安装和使用指南
Requests库安装与使用指南Requests是一个功能强大且易于使用的PythonHTTP库,广泛应用于发送各种HTTP请求,如GET、POST等。以下内容将详细介绍Requests库的安装和使用方法,帮助您高效地在Python中进行HTTP操作。️安装Requests库要使用Requests库,首先需要确保已安装pip工具。然后,在终端或命令行中运行以下命令进行安装:pipinstallre
- 用Python在Excel工作表中创建数据透视图
数据透视图是基于数据透视表创建的Excel图标,它能够帮助我们从复杂的数据集中提炼出有价值的信息,提供直观且易于理解的数据视图。对于需要频繁更新或处理大量数据集的人员以及任何依赖数据做出决策的人来说,用Python在Excel中创建数据透视图能够根据最新的数据快速调整和生成新的分析图表,从而提高工作效率并增强数据分析的灵活性。本文将介绍如何使用Python在Excel工作表中创建数据透视图。用Py
- Go Ebiten小游戏开发:贪吃蛇
RedJACK~
小游戏开发Go语言golang开发语言后端
贪吃蛇是一款经典的小游戏,玩法简单却充满乐趣。本文将介绍如何使用Go语言和Ebiten游戏引擎开发一个简单的贪吃蛇游戏。通过这个项目,你可以学习到游戏开发的基本流程、Ebiten的使用方法以及如何用Go实现游戏逻辑。项目简介贪吃蛇的核心玩法是控制一条蛇在网格中移动,吃掉随机生成的食物,每吃一个食物蛇身会变长,同时得分增加。如果蛇撞到墙壁或自己的身体,游戏结束。本项目使用Go语言和Ebiten游戏引
- C/C++三方库编译构建
harmonyos
课程简介本课程是【HarmonyOSTechTalk】的第19课。本次交流聚焦于C/C++三方库在HarmonyOS开发中的应用。首先是适配HarmonyOS工具链,这是将开源三方库融入鸿蒙生态的关键步骤,确保其兼容性与稳定性。DevEcoStudio则是构建的得力助手,可用于打造自定义三方库,满足特定开发需求。在Native工程里使用这些三方库,能拓展功能、提升效率。通过本次课程学习,开发者能够
- SGCN模型详解及代码复现
呆头鹅AI工作室
深度学习算法详解及代码复现深度学习人工智能自然语言处理神经网络python
模型背景SGCN模型源于2018年ICDM会议的一项开创性研究,旨在解决传统图卷积网络(GCNs)在处理签名图时面临的挑战。签名图包含正负链接,反映实体间复杂的相互作用,如社交媒体中的点赞和屏蔽关系。SGCN通过巧妙结合平衡理论和图卷积操作,实现了对正负链接的有效处理,在节点表示学习任务中展现出卓越性能,为社交网络分析、链接预测和社区检测等领域提供了新思路。核心思想SGCN模型的核心思想在于其创新
- pip工具安装第三方库
nfenghklibra
pippython
使用pip+cmd引入第三方库pip是Python包管理工具,提供了对Python包的查找、下载、安装、卸载的功能。注意:pip已内置于Python3.4和2.7及以上版本,其他版本需另行安装常规命令:pipinstall安装第三方库的库名(以json为例)pipinstalljson指定版本号:pipinstall库名==库的版本号pipinstalljieba==0.42.1卸载库:pipun
- python 词云示例
布道天下
python
python词云示例以2021年中央1号文件和政府工作报告文件为例,输出50个关键词。#testPython.pyimportjiebaimportwordclouddefoutputWordCloud(text,outPngName):#配置词云对象参数temp=wordcloud.WordCloud(width=1000,height=1000,font_path="msyh.ttc",max
- requests库的安装和使用指南
Requests库安装与使用指南Requests是一个简洁且功能强大的Python库,用于发送HTTP请求。它广泛应用于数据采集、API调用等场景。本文将详细介绍Requests库的安装与基本使用方法,并通过实例和图表帮助您快速掌握其核心功能。目录安装Requests库导入Requests库发送GET请求发送POST请求添加Headers处理响应处理JSON响应异常处理附加参数会话管理文件下载工作
- 【Python基础 字典】汽车限行
学Python的小趴菜
python
最近在准备期末机考,看实验课的代码,发现有提升效率的空间,就改了改测评过了。这个效率提升是砍掉了循环结构判断车牌号末尾数字的奇偶,改用纯数学方法(提取数字判断奇偶)任务描述为缓解城市交通压力,武汉市交管局对于长江一桥及江汉一桥实行限行,规定如下:
- Python小项目:利用U-net完成细胞图像分割
利用U-Net完成细胞图像分割的详细指南在生物医学领域,细胞图像分割是一个关键步骤,能够帮助研究人员分析细胞结构和功能。U-Net作为一种强大的卷积神经网络结构,广泛应用于医学图像分割任务。本文将详细介绍如何利用U-Net完成细胞图像分割项目,涵盖从数据准备到模型部署的各个步骤。项目步骤概览数据准备数据预处理构建U-Net模型训练模型模型评估图像分割结果可视化调优和优化部署和应用1.数据准备收集数
- json相关内容(python)
大哥喝阔落
jsonpython开发语言
JSON(JavaScriptObjectNotation)是一种轻量级的数据交换格式,易于人阅读和编写,也易于机器解析和生成。Python提供了json模块来处理JSON数据。以下是关于Python中JSON的详细内容:1.导入json模块importjson2.将Python对象转换为JSON字符串使用json.dumps()函数可以将Python对象(如字典、列表、字符串、数字等)转换为JS
- 大数据学习笔记——zookeeper在hadoop集群中的作用
鹅鹅鹅呢
javahadoop大数据学习tcp/iptomcat
zookeeper主要是用来搭建高可用的Hadoop集群,即HighAvailability,简称(HA)测试中集群是可以不需要高可用的,即使用一个namenode即可。但是在生产环境中为了提高集群的可靠性,需要增加一个namenode备用,当active的namenode挂了之后,系统会启动standby的namenode。这就需要zookeeper监控namenode的状态。
- PyEcharts 基本图表之词云图
开不开心少年
头哥题目python开发语言
第1关:WordCloud:词云图任务描述本关任务:利用所学知识,按要求自行绘制一个词云图。相关知识为了完成本关任务,你需要掌握:1.Python的基本语法,2.PyEcharts词云图的相关内容。编程要求根据以上介绍,在右侧编辑器补充代码,使用给定数据绘制一个词云图,要求:系列名称设置为空,数据项为data,单词字体大小范围设置为20到100,词云图轮廓设置为全局变量中的SymbolType.D
- Nginx Proxy Manager 反代本地服务502错误——基于 1panel 部署遇到的问题解决方案
nginx后端python
参考:NginxProxyManager反代本地服务502错误我的需求如下:我有一个需求:我有一台云服务器,ip地址为114.55.xxx.xxx然后在这个机器上部署了一个python服务http://114.55.xxx.xxx:8086我需要实现一个功能:部署一个nginx当我访问云服务器的80端口的时候,可以帮我反向代理访问python服务,我应该如何设置nginx呢?给我对应的配置文件配置
- 基于Tkinter和Canvas实现PCB产品的Map分布展示
卤蛋叔叔
TkinterCanvasMappython
本文是基于SEMI的G85文件(文件格式类型为XML),和Python的Tkinter和Canvas模块实现PCB/SUB(芯片基板)类型的产品的检测缺陷的Map分布展示,第一部分主要对程序的页面进行展示1、Lot_Map叠合的页面主要分为两个页面图,通过点击获取文件按钮(获取的文件格式为固定的G85文件,示例如图2),获取指定格式路径下的文件后,左边页面:点击获取MergeMap明细,可获取到所
- Python小工具:利用ffmpy3库3秒钟将视频转换为音频
Python知识圈
python
作者|pk哥来源公众号|Python知识圈(ID:PythonCircle)最近,有读者微信上私聊我,想让我写一篇视频批量转换成音频的文章,我答应了,周末宅家里把这个小工具做出来了。这样,对于有些视频学习文件,我们可以批量转换成音频文件,学习方式更多样化了。之前也用过ffmpeg处理视频文件。ffmpeg这个程序处理视频是好用,但是有没有更轻便的呢?可以不下载这个程序吗?还真有,Python里有f
- 开源 多模态 大模型架构深度分析 2024
AI大模型 lose and dream
开源架构学习langchainprompt人工智能开发语言
1.典型开源多模态大模型(1)KOSMOS-2KOSMOS-2是微软亚洲研究院在KOSMOS-1模型的基础上开发的多模态大模型。其中,KOSMOS-1是在大规模多模态数据集上重头训练的,该模型具有类似GPT-4的多模态能力,可以感知一般的感官模态,在上下文中学习(即少样本学习)并能够遵循语音指示(即零样本学习)。KOSMOS-2采用与KOSMOS-1相同的模型架构和训练目标对模型进行训练,并在此基
- 应急救援路径规划中的蚁群算法与路径评价研究【附代码】
拉勾科研工作室
算法
数据科学与大数据专业|数据分析与模型构建|数据驱动决策✨专业领域:数据挖掘与清洗大数据处理与存储技术机器学习与深度学习模型数据可视化与报告生成分布式计算与云计算数据安全与隐私保护擅长工具:Python/R/Matlab数据分析与建模Hadoop/Spark大数据处理平台SQL数据库管理与优化Tableau/PowerBI数据可视化工具TensorFlow/PyTorch深度学习框架✅具体问题可以私
- 【列表复制】详解python中list列表复制的几种方法(赋值、切片、copy(),deepcopy())
有梦想的程序星空
Python开发教程python开发语言
在Python编程领域,列表是一种极为常用的数据结构,用于存储多个元素的有序集合。当涉及到对列表进行复制操作时,浅拷贝和深拷贝是两种重要的概念与技术手段,它们在处理列表数据的过程中有着截然不同的行为和影响,深刻理解二者的差异与应用场景对于编写高效、准确且健壮的Python代码至关重要。1、浅拷贝和深拷贝浅拷贝复制指向某个对象的地址(指针),而不复制对象本身,新对象和原对象共享同一内存。深拷贝会额外
- Hugging Face Transformers 库学习提纲
做个天秤座的程序猿
HuggingFaceTransformers学习transformerpython
文章目录前言一、[基础概念](https://blog.csdn.net/kljyrx/article/details/139984730)二、[环境准备](https://blog.csdn.net/kljyrx/article/details/140006571)三、库的基本使用四、高级应用五、实践案例六、生态系统和工具七、社区与资源八、进阶学习总结前言HuggingFaceTransform
- 2024年大数据最全【ES专题】ElasticSearch集群架构剖析_es集群
kenzsoft
程序员大数据elasticsearch架构
IngestNode:数据前置处理转换节点,支持pipeline管道设置,可以使用ingest对数据进行过滤、转换等操作MachineLearningNode:负责跑机器学习的Job,用来做异常检测TribeNode:TribeNode连接到不同的Elasticsearch集群,并且支持将这些集群当成一个单独的集群处理以下是一个多集群业务架构图:1.2.1.1MasterNode主节点的功能Mas
- python3安装clickhouse_sqlalchemy(greenlet) 失败
安装clickhouse_sqlalchemy时,可能会遇到依赖问题,特别是greenlet模块的安装问题。以下是详细的解决方案,帮助您顺利完成安装过程。常见问题与解决方案1.升级pip确保您的pip版本是最新的,这有助于避免由于旧版本导致的兼容性问题。pipinstall--upgradepip解释:使用pipinstall--upgradepip命令将pip升级到最新版本,以确保能够安装最新的
- python(类和对象之类函数和静态函数)
huo_1214
类函数和静态函数#-*-coding:utf-8-*-#类函数和静态函数classPeople(object):#类变量total=0def__init__(self,name,age):#调用父类的初始化函数super(People,self).__init__()#初始化当前类对象的一些属性self.name=nameself.age=age#对象函数,只能由对象调用defeat(self):
- 知乎高赞!BAT大牛的大数据学习之路!
数据工程师金牛
大数据大数据人工智能机器学习数据挖掘数据分析
前几天,网易云音乐公布了一份年度音乐总结。让我惊叹的是在这个大数据时代底下,比起我们自己,大数据似乎更懂得我们。如果科技更进一步,就像《奇葩说》中一集辩题里所说的,它有可能可以帮我们匹配到那个灵魂相契的人。有人觉得这个现象很恐怖,但在我看来,人的恐惧是源自于未知。如果能顺应着时代的步伐,一起向前,对它了解再加深一点,也许我们就会爱上它。在这里相信有许多想要学习大数据的同学,大家可以+下大数据学习裙
- python程序设计期末大作业,python大作业代码100行
chatgpt001
人工智能
大家好,小编来为大家解答以下问题,python期末大作业代码200行带批注,python程序设计期末大作业,今天让我们一起来看看吧!#题目:利用Python实现一个计算器,可以计算小数复数等importredefcalculator(string):#去除括号函数defget_grouping(string):flag=Falseret=re.findall('\(([^()]+)\)',stri
- HarmonyOS Next V2 状态管理 AppStorageV2 和 PersistenceV2
harmonyos
HarmonyOSNextV2状态管理AppStorageV2和PersistenceV2前言在HarmonyOS应用开发过程中,我们已经学习过了不少关于状态管理相关的技术,如@ObservedV2装饰器和@Trace装饰器:类属性变化观测@ComponentV2装饰器:自定义组件@Local装饰器:组件内部状态@Param:组件外部输入@Once:初始化同步一次@Event装饰器:组件输出@Mo
- 华为OD机试E卷 - 单词接龙(Java & Python& JS & C++ & C )
算法大师
最新华为OD机试华为odjavapythonjavascriptc++C
最新华为OD机试真题目录:点击查看目录华为OD面试真题精选:点击立即查看题目描述单词接龙的规则是:可用于接龙的单词首字母必须要前一个单词的尾字母相同;当存在多个首字母相同的单词时,取长度最长的单词,如果长度也相等,则取字典序最小的单词;已经参与接龙的单词不能重复使用。现给定一组全部由小写字母组成单词数组,并指定其中的一个单词作为起始单词,进行单词接龙,请输出最长的单词串,单词串是单词拼接而成,中间
- JAVA基础
灵静志远
位运算加载Date字符串池覆盖
一、类的初始化顺序
1 (静态变量,静态代码块)-->(变量,初始化块)--> 构造器
同一括号里的,根据它们在程序中的顺序来决定。上面所述是同一类中。如果是继承的情况,那就在父类到子类交替初始化。
二、String
1 String a = "abc";
JAVA虚拟机首先在字符串池中查找是否已经存在了值为"abc"的对象,根
- keepalived实现redis主从高可用
bylijinnan
redis
方案说明
两台机器(称为A和B),以统一的VIP对外提供服务
1.正常情况下,A和B都启动,B会把A的数据同步过来(B is slave of A)
2.当A挂了后,VIP漂移到B;B的keepalived 通知redis 执行:slaveof no one,由B提供服务
3.当A起来后,VIP不切换,仍在B上面;而A的keepalived 通知redis 执行slaveof B,开始
- java文件操作大全
0624chenhong
java
最近在博客园看到一篇比较全面的文件操作文章,转过来留着。
http://www.cnblogs.com/zhuocheng/archive/2011/12/12/2285290.html
转自http://blog.sina.com.cn/s/blog_4a9f789a0100ik3p.html
一.获得控制台用户输入的信息
&nbs
- android学习任务
不懂事的小屁孩
工作
任务
完成情况 搞清楚带箭头的pupupwindows和不带的使用 已完成 熟练使用pupupwindows和alertdialog,并搞清楚两者的区别 已完成 熟练使用android的线程handler,并敲示例代码 进行中 了解游戏2048的流程,并完成其代码工作 进行中-差几个actionbar 研究一下android的动画效果,写一个实例 已完成 复习fragem
- zoom.js
换个号韩国红果果
oom
它的基于bootstrap 的
https://raw.github.com/twbs/bootstrap/master/js/transition.js transition.js模块引用顺序
<link rel="stylesheet" href="style/zoom.css">
<script src=&q
- 详解Oracle云操作系统Solaris 11.2
蓝儿唯美
Solaris
当Oracle发布Solaris 11时,它将自己的操作系统称为第一个面向云的操作系统。Oracle在发布Solaris 11.2时继续它以云为中心的基调。但是,这些说法没有告诉我们为什么Solaris是配得上云的。幸好,我们不需要等太久。Solaris11.2有4个重要的技术可以在一个有效的云实现中发挥重要作用:OpenStack、内核域、统一存档(UA)和弹性虚拟交换(EVS)。
- spring学习——springmvc(一)
a-john
springMVC
Spring MVC基于模型-视图-控制器(Model-View-Controller,MVC)实现,能够帮助我们构建像Spring框架那样灵活和松耦合的Web应用程序。
1,跟踪Spring MVC的请求
请求的第一站是Spring的DispatcherServlet。与大多数基于Java的Web框架一样,Spring MVC所有的请求都会通过一个前端控制器Servlet。前
- hdu4342 History repeat itself-------多校联合五
aijuans
数论
水题就不多说什么了。
#include<iostream>#include<cstdlib>#include<stdio.h>#define ll __int64using namespace std;int main(){ int t; ll n; scanf("%d",&t); while(t--)
- EJB和javabean的区别
asia007
beanejb
EJB不是一般的JavaBean,EJB是企业级JavaBean,EJB一共分为3种,实体Bean,消息Bean,会话Bean,书写EJB是需要遵循一定的规范的,具体规范你可以参考相关的资料.另外,要运行EJB,你需要相应的EJB容器,比如Weblogic,Jboss等,而JavaBean不需要,只需要安装Tomcat就可以了
1.EJB用于服务端应用开发, 而JavaBeans
- Struts的action和Result总结
百合不是茶
strutsAction配置Result配置
一:Action的配置详解:
下面是一个Struts中一个空的Struts.xml的配置文件
<?xml version="1.0" encoding="UTF-8" ?>
<!DOCTYPE struts PUBLIC
&quo
- 如何带好自已的团队
bijian1013
项目管理团队管理团队
在网上看到博客"
怎么才能让团队成员好好干活"的评论,觉得写的比较好。 原文如下: 我做团队管理有几年了吧,我和你分享一下我认为带好团队的几点:
1.诚信
对团队内成员,无论是技术研究、交流、问题探讨,要尽可能的保持一种诚信的态度,用心去做好,你的团队会感觉得到。 2.努力提
- Java代码混淆工具
sunjing
ProGuard
Open Source Obfuscators
ProGuard
http://java-source.net/open-source/obfuscators/proguardProGuard is a free Java class file shrinker and obfuscator. It can detect and remove unused classes, fields, m
- 【Redis三】基于Redis sentinel的自动failover主从复制
bit1129
redis
在第二篇中使用2.8.17搭建了主从复制,但是它存在Master单点问题,为了解决这个问题,Redis从2.6开始引入sentinel,用于监控和管理Redis的主从复制环境,进行自动failover,即Master挂了后,sentinel自动从从服务器选出一个Master使主从复制集群仍然可以工作,如果Master醒来再次加入集群,只能以从服务器的形式工作。
什么是Sentine
- 使用代理实现Hibernate Dao层自动事务
白糖_
DAOspringAOP框架Hibernate
都说spring利用AOP实现自动事务处理机制非常好,但在只有hibernate这个框架情况下,我们开启session、管理事务就往往很麻烦。
public void save(Object obj){
Session session = this.getSession();
Transaction tran = session.beginTransaction();
try
- maven3实战读书笔记
braveCS
maven3
Maven简介
是什么?
Is a software project management and comprehension tool.项目管理工具
是基于POM概念(工程对象模型)
[设计重复、编码重复、文档重复、构建重复,maven最大化消除了构建的重复]
[与XP:简单、交流与反馈;测试驱动开发、十分钟构建、持续集成、富有信息的工作区]
功能:
- 编程之美-子数组的最大乘积
bylijinnan
编程之美
public class MaxProduct {
/**
* 编程之美 子数组的最大乘积
* 题目: 给定一个长度为N的整数数组,只允许使用乘法,不能用除法,计算任意N-1个数的组合中乘积中最大的一组,并写出算法的时间复杂度。
* 以下程序对应书上两种方法,求得“乘积中最大的一组”的乘积——都是有溢出的可能的。
* 但按题目的意思,是要求得这个子数组,而不
- 读书笔记-2
chengxuyuancsdn
读书笔记
1、反射
2、oracle年-月-日 时-分-秒
3、oracle创建有参、无参函数
4、oracle行转列
5、Struts2拦截器
6、Filter过滤器(web.xml)
1、反射
(1)检查类的结构
在java.lang.reflect包里有3个类Field,Method,Constructor分别用于描述类的域、方法和构造器。
2、oracle年月日时分秒
s
- [求学与房地产]慎重选择IT培训学校
comsci
it
关于培训学校的教学和教师的问题,我们就不讨论了,我主要关心的是这个问题
培训学校的教学楼和宿舍的环境和稳定性问题
我们大家都知道,房子是一个比较昂贵的东西,特别是那种能够当教室的房子...
&nb
- RMAN配置中通道(CHANNEL)相关参数 PARALLELISM 、FILESPERSET的关系
daizj
oraclermanfilespersetPARALLELISM
RMAN配置中通道(CHANNEL)相关参数 PARALLELISM 、FILESPERSET的关系 转
PARALLELISM ---
我们还可以通过parallelism参数来指定同时"自动"创建多少个通道:
RMAN > configure device type disk parallelism 3 ;
表示启动三个通道,可以加快备份恢复的速度。
- 简单排序:冒泡排序
dieslrae
冒泡排序
public void bubbleSort(int[] array){
for(int i=1;i<array.length;i++){
for(int k=0;k<array.length-i;k++){
if(array[k] > array[k+1]){
- 初二上学期难记单词三
dcj3sjt126com
sciet
concert 音乐会
tonight 今晚
famous 有名的;著名的
song 歌曲
thousand 千
accident 事故;灾难
careless 粗心的,大意的
break 折断;断裂;破碎
heart 心(脏)
happen 偶尔发生,碰巧
tourist 旅游者;观光者
science (自然)科学
marry 结婚
subject 题目;
- I.安装Memcahce 1. 安装依赖包libevent Memcache需要安装libevent,所以安装前可能需要执行 Shell代码 收藏代码
dcj3sjt126com
redis
wget http://download.redis.io/redis-stable.tar.gz
tar xvzf redis-stable.tar.gz
cd redis-stable
make
前面3步应该没有问题,主要的问题是执行make的时候,出现了异常。
异常一:
make[2]: cc: Command not found
异常原因:没有安装g
- 并发容器
shuizhaosi888
并发容器
通过并发容器来改善同步容器的性能,同步容器将所有对容器状态的访问都串行化,来实现线程安全,这种方式严重降低并发性,当多个线程访问时,吞吐量严重降低。
并发容器ConcurrentHashMap
替代同步基于散列的Map,通过Lock控制。
&nb
- Spring Security(12)——Remember-Me功能
234390216
Spring SecurityRemember Me记住我
Remember-Me功能
目录
1.1 概述
1.2 基于简单加密token的方法
1.3 基于持久化token的方法
1.4 Remember-Me相关接口和实现
- 位运算
焦志广
位运算
一、位运算符C语言提供了六种位运算符:
& 按位与
| 按位或
^ 按位异或
~ 取反
<< 左移
>> 右移
1. 按位与运算 按位与运算符"&"是双目运算符。其功能是参与运算的两数各对应的二进位相与。只有对应的两个二进位均为1时,结果位才为1 ,否则为0。参与运算的数以补码方式出现。
例如:9&am
- nodejs 数据库连接 mongodb mysql
liguangsong
mongodbmysqlnode数据库连接
1.mysql 连接
package.json中dependencies加入
"mysql":"~2.7.0"
执行 npm install
在config 下创建文件 database.js
- java动态编译
olive6615
javaHotSpotjvm动态编译
在HotSpot虚拟机中,有两个技术是至关重要的,即动态编译(Dynamic compilation)和Profiling。
HotSpot是如何动态编译Javad的bytecode呢?Java bytecode是以解释方式被load到虚拟机的。HotSpot里有一个运行监视器,即Profile Monitor,专门监视
- Storm0.9.5的集群部署配置优化
roadrunners
优化storm.yaml
nimbus结点配置(storm.yaml)信息:
# Licensed to the Apache Software Foundation (ASF) under one
# or more contributor license agreements. See the NOTICE file
# distributed with this work for additional inf
- 101个MySQL 的调节和优化的提示
tomcat_oracle
mysql
1. 拥有足够的物理内存来把整个InnoDB文件加载到内存中——在内存中访问文件时的速度要比在硬盘中访问时快的多。 2. 不惜一切代价避免使用Swap交换分区 – 交换时是从硬盘读取的,它的速度很慢。 3. 使用电池供电的RAM(注:RAM即随机存储器)。 4. 使用高级的RAID(注:Redundant Arrays of Inexpensive Disks,即磁盘阵列
- zoj 3829 Known Notation(贪心)
阿尔萨斯
ZOJ
题目链接:zoj 3829 Known Notation
题目大意:给定一个不完整的后缀表达式,要求有2种不同操作,用尽量少的操作使得表达式完整。
解题思路:贪心,数字的个数要要保证比∗的个数多1,不够的话优先补在开头是最优的。然后遍历一遍字符串,碰到数字+1,碰到∗-1,保证数字的个数大于等1,如果不够减的话,可以和最后面的一个数字交换位置(用栈维护十分方便),因为添加和交换代价都是1