lasso回归_生存分析之lasso回归代码
 点击上方“小梦游仙境” 带你去看小星星
点击上方“小梦游仙境” 带你去看小星星
代码来自老大github:https://github.com/jmzeng1314/tcga_example中的step05-lasso
这篇仅仅是代码,图片的解析后面学习过再继续记录呢~
 获得生存资料和表达矩阵数据
获得生存资料和表达矩阵数据
rm(list=ls())
options(stringsAsFactors = F)
Rdata_dir='../Rdata/'
Figure_dir='../figures/'
# 加载上一步从RTCGA.miRNASeq包里面提取miRNA表达矩阵和对应的样本临床信息。
load( file =
file.path(Rdata_dir,'TCGA-KIRC-miRNA-example.Rdata')
)
dim(expr)
dim(meta)
# 可以看到是 537个病人,但是有593个样本,每个样本有 552个miRNA信息。
# 当然,这个数据集可以下载原始测序数据进行重新比对,可以拿到更多的miRNA信息
# 这里需要解析TCGA数据库的ID规律,来判断样本归类问题。
group_list=ifelse(as.numeric(substr(colnames(expr),14,15)) 10,'tumor','normal')
table(group_list)
exprSet=na.omit(expr)
table(group_list)
exprSet=na.omit(expr)
dim(exprSet)
load( file =
file.path(Rdata_dir,'TCGA-KIRC-miRNA-survival_input.Rdata')
)
dim(exprSet) ## remove the nomral
head(phe)
exprSet[1:4,1:4]
head(colnames(exprSet))
head(phe$ID)
## 必须保证生存资料和表达矩阵,两者一致
all(substring(colnames(exprSet),1,12)==phe$ID)
 lasso回归系列代码
lasso回归系列代码
library(survival)
library(survminer)
library(lars)
library(glmnet)
x=t(log2(exprSet+1))
y=phe$event
> model_lasso "binomial", nlambda=50, alpha=1)
> print(model_lasso)
Call: glmnet(x = x, y = y, family = "binomial", alpha = 1, nlambda = 50)
Df %Dev Lambda
1 0 0.00000 0.127800
2 3 0.01364 0.116300
3 3 0.02631 0.105900
4 4 0.03873 0.096400
5 4 0.04984 0.087750
6 6 0.06510 0.079880
#此处省略中间
45 234 0.88730 0.002045
46 232 0.89760 0.001861
47 237 0.90690 0.001694
48 239 0.91530 0.001542
49 241 0.92300 0.001404
50 244 0.92990 0.001278
上面的结果解释如下:
列%Dev代表了由模型解释的残差的比例,对于线性模型来说就是模型拟合的R^2(R-squred)。
它在0和1之间,越接近1说明模型的表现越好,
如果是0,说明模型的预测结果还不如直接把因变量的均值作为预测值来的有效。
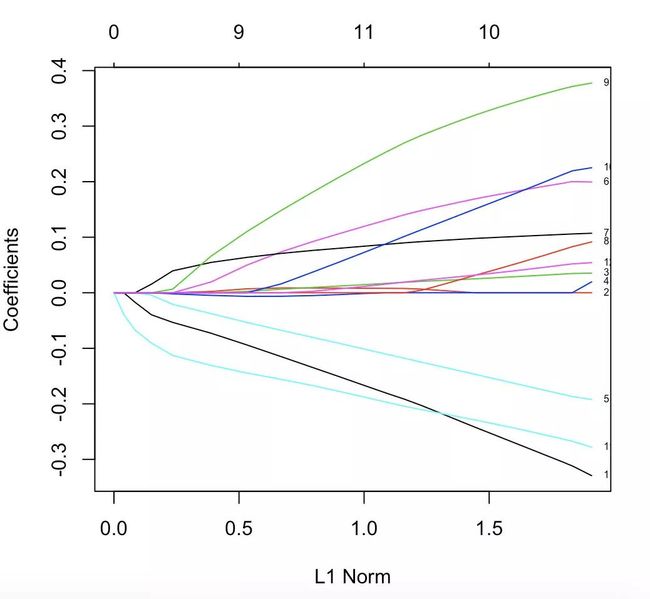
plot(model_lasso, xvar = "norm", label = TRUE)

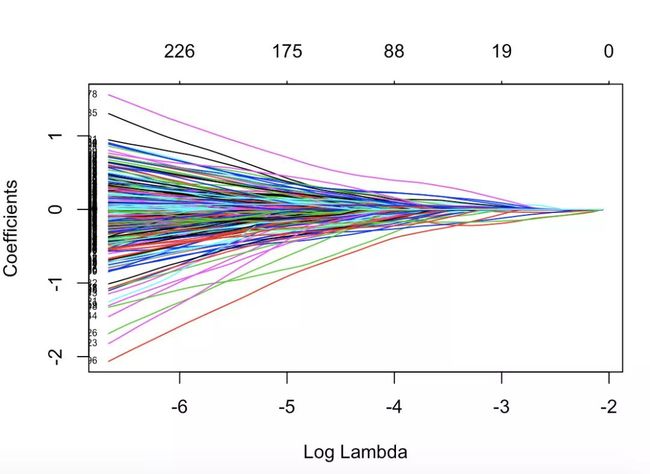
plot(model_lasso, xvar="lambda", label=TRUE)
cv_fit 1, nlambda = 1000)
plot(cv_fit)
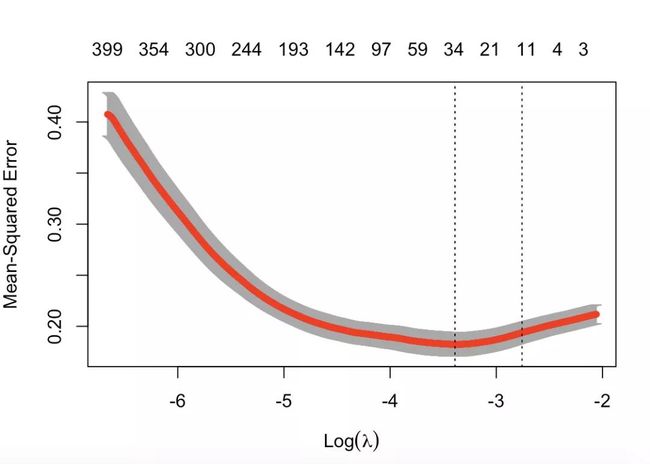
# 上面两条虚线分别指示了两个特殊的λ值:
c(cv_fit$lambda.min,cv_fit$lambda.1se)
[1] 0.03372476 0.06341954
model_lasso1 1, lambda=cv_fit$lambda.1se)
lasso.prob re=cbind(y ,lasso.prob)
dat=as.data.frame(re[,1:2])
colnames(dat)=c('event','prob')
dat$event=as.factor(dat$event)
library(ggpubr)
p "event", y = "prob",
color = "event", palette = "jco",
add = "jitter")
# Add p-value
p + stat_compare_means()

library(ROCR)
library(glmnet)
library(caret)
# calculate probabilities for TPR/FPR for predictions
pred 2], re[,1])
perf "tpr","fpr")
performance(pred,"auc") # shows calculated AUC for model
plot(perf,colorize=FALSE, col="black") # plot ROC curve
lines(c(0,1),c(0,1),col = "gray", lty = 4 )

> fit 1, lambda=cv_fit$lambda.1se)
> head(fit$beta)
6 x 1 sparse Matrix of class "dgCMatrix"
s0
hsa-let-7a-1 .
hsa-let-7a-2 .
hsa-let-7a-3 .
hsa-let-7b .
hsa-let-7c .
hsa-let-7d .
choose_gene=rownames(fit$beta)[as.numeric(fit$beta)!=0]
length(choose_gene)
myexpr=x[,choose_gene]
mysurv=phe[,c("days","event")]
mysurv$days[mysurv$days1] = 1
# 详细代码参见这个网站https://github.com/jeffwong/glmnet/blob/master/R/coxnet.R#
fit family = "cox")
用包自带的函数画图
plot(fit, xvar="lambda", label = TRUE)
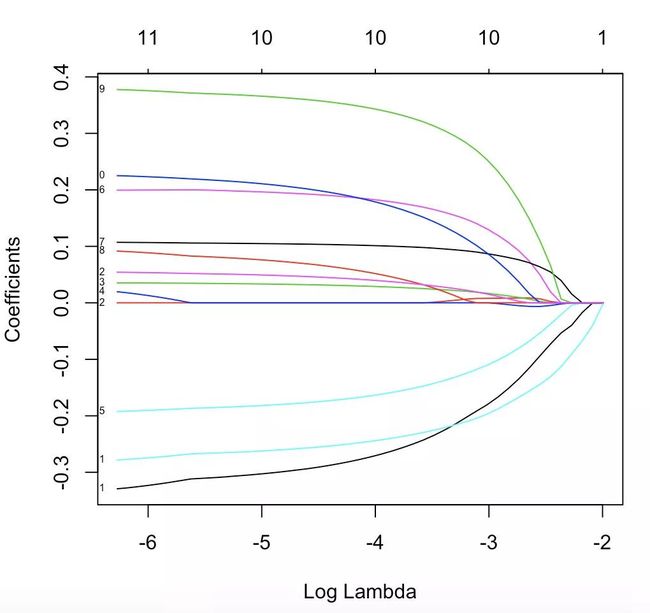
plot(fit, label = TRUE)