蛋白质分子结构设计
paper read 1
Created by: 银晗 张
Created time: May 27, 2023 3:47 PM
Tags: Product
- 补充了解蛋白质的生物学知识
- 学习一下Diffusion的原理
Method & Innovations
- Framework
Summary:
- first deep learning models to perform antibody sequence-structure design by considering the 3D structures of the antigen
- design protein sequences and coordinates & side-chain orientations , firstly achieve atomic-resolution antibody design and is equivariant to rotation and translationw
- applied to antibody design tasks sequence-structure co-design, fix-backbone CDR design, and antibody optimization
Prospose Method: 基于扩散的生成模型联合采样抗体CDR序列和结构
- CDR序列及其结构的联合分布直接依赖于抗原结构, 所以我们的任务是给定一个由抗原和抗体框架组成的蛋白质复合物作为输入,得到CDRs的结构
- Differences to previous works
Traditional Computational Antibody Design Problems:
- the search space of CDRs is vast , L squences may have 20^L
- time-consuming and local optima
Generative model challenges :
- how to model the intrinsic relation between CDR sequences and 3D structures
- how to model the distribution of CDRs conditional on the rest of the antibody sequence
- the model should be explicitly conditional on the 3D structures of the antigen and generate CDRs that fit the antigen structure in the 3D space
- model should be able to consider both the position and orientation of amino acids
- instead of de novo design, model should be applicable to another realistic scenario: optimizing a particular antibody to increase the binding affinity to the antigen
Related Diffusion-Based Generative Models
- the sequence-based methods can only generate new antibodies based on previously observed
antibodies but can hardly generate antibodies for specific antigen structures - protein structure pretidion algorithms : MSAs、AlphaFold2
- diffusion model : denosing with prior distrubtion、molecular 3D structure
Differences
M o d e l S t e p s : Model Steps: ModelSteps:
- 用任意序列、位置和方向初始化CDR。扩散模型首先聚集了来自抗原和抗体框架的信息
- 迭代地更新cdr上每个氨基酸的氨基酸类型、位置和方向(侧链的方向)
- 我们基于预测的方向,使用侧链填充算法在原子级重建CDR结构
- What insights would the proposed approach bring?
SO(3) Denosing for Amino Acid Orientations:
S : coordinates , X: amino acid types, O: orientations
- 各向同性的高斯分布,改变旋转角度
- 神经网络用于方向去噪和输出去噪的方向矩阵
- 目标函数是真实和预测的方向矩阵之间的差异内积
Diffusion For C a C_a Ca Coordinates :
- 坐标是一个正态分布
- 变化的学习率
- 神经网络用于预测高斯分布的噪声
- 目标函数是生成的分布和初始先验分布的MSE
Migrate Markov chains
-
A diffusion probabilistic model defines two Markov chains of diffusion processes
- The forward diffusion process gradually adds noise to the data until the data distribution approximately reaches the prior distribution
- The generative diffusion process starts from the prior distribution and iteratively
transforms it to the desired distribution.
-
Use
Multinomial Distributioninstead ofGaussian distributionin Diffusion Model
Processes:
-
MLP embeding: encodes the information of amino acid types, torsional angles, and 3D coordinates of all the heavy atoms . The pairwise embedding MLP encodes the Euclidean distances and dihedral angles between amino acid i and j to feature vectors zij, use IPA(to transform ∗ ∗ ∗ e i ∗ ∗ ∗ ***e_i*** ∗∗∗ei∗∗∗ and ∗ ∗ ∗ z i j ∗ ∗ ***z_{ij}** ∗∗∗zij∗∗ into hidden representations ∗ ∗ ∗ h i ∗ ∗ ***h_i** ∗∗∗hi∗∗) to represent itself and environment
-
Denoise: the representations are fed to three different MLPs to denoise the amino acid types, 3D positions, and orientations of the CDR,respectively
-
预测局部坐标系中的坐标偏差,并将其投影到全局坐标系中,可以确保预测的等方差,因为当整个三维结构以特定的角度旋转时,坐标偏差也以相同的角度旋转。
-
偏差表示:
-
将向量转换为旋转矩阵 M j ∈ S O ( 3 ) M_j∈SO (3) Mj∈SO(3)右向乘以方向,为下一步生成步骤产生一个新的平均方向: O j t − 1 ← O j t M j O^{t−1}_j←O^t_jM_j Ojt−1←OjtMj。
-
所提出的网络与整体结构的旋转和平移是等变的:
-
-
sample algorithm:
- 20 types amino acids distrubution : s j T ∼ U n i f o r m ( 20 ) s^T_j∼ Uniform(20) sjT∼Uniform(20)
- C α C_α Cα positions from the standard normal distribution: ∗ ∗ x j T ∼ N ( 0 , I 3 ) **x^T_j ∼ N (0, I3) ∗∗xjT∼N(0,I3), side-chain C β C_β Cβ
- orientations from the uniform distribution over SO(3): ∗ ∗ O j T ∼ U n i f o r m ( S O ( 3 ) ) ∗ ∗ **O^T_j∼ Uniform(SO(3))** ∗∗OjT∼Uniform(SO(3))∗∗
DiffAb的实验:
DiffAb Experiment
Details
Antigen-Specific Antibody Design and Optimization with Diffusion-Based Generative Models for Protein Structures
Domain words:
- antigen, antibody : 抗体、抗原
- complementarity-determining regions (CDR):互补性结构区域
- amino acids: 氨基酸
- molecular , atom : 分子、原子
SO(3)是三维旋转群的代数结构,表示在三维空间中的旋转操作。在蛋白质中,SO(3)通常用于描述氨基酸侧链的取向。通过SO(3)变换,可以将一个氨基酸侧链的取向转换为另一个取向,从而对氨基酸侧链的构象空间进行建模和分析。
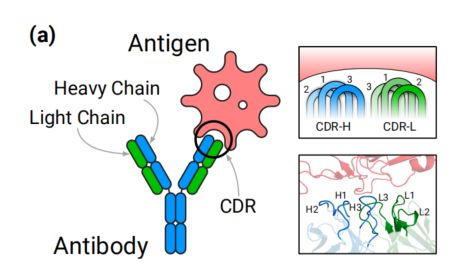
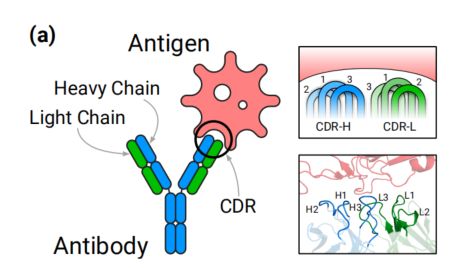
抗体是免疫系统蛋白质,通过与病毒和细菌等特定抗原结合来保护宿主。抗体与抗原之间的结合主要由抗体的互补决定区(CDR)决定。在这项工作中,我们开发了一个深度生成模型,基于扩散概率模型和等变神经网络,共同模拟CDR的序列和结构。我们的方法是第一个明确针对特定抗原结构生成抗体的深度学习方法,并且是早期蛋白质结构扩散概率模型之一。
该模型能够进行序列-结构协同设计、给定骨架结构的序列设计和抗体优化。
我们进行了广泛的实验,评估了设计抗体的序列和结构的质量。我们发现,我们的模型在生物物理能量函数和其他蛋白质设计指标衡量的结合亲和力方面能够产生有竞争力的结果。
Background
- the structure of antibody-antigen
Target: to design effective therapeutic antibody structure
Traditional Problems:
- the search space of CDRs is vast , L squences may have 20^L
- time-consuming and local optima
Generative model challenges :
- how to model the intrinsic relation between CDR sequences and 3D structures
- how to model the distribution of CDRs conditional on the rest of the antibody sequence
- the model should be explicitly conditional on the 3D structures of the antigen and generate CDRs that fit the antigen structure in the 3D space
- model should be able to consider both the position and orientation of amino acids
- instead of de novo design, model should be applicable to another realistic scenario: optimizing a particular antibody to increase the binding affinity to the antigen
- the sequence-based methods can only generate new antibodies based on previously observed
antibodies but can hardly generate antibodies for specific antigen structures - protein structure pretidion algorithms : MSAs、AlphaFold2
- diffusion model : 先验分布去噪、molecular 3D structure
Work
Prospose Method: 基于扩散的生成模型联合采样抗体CDR序列和结构
- CDR序列及其结构的联合分布直接依赖于抗原结构, 所以我们的任务是给定一个由抗原和抗体框架组成的蛋白质复合物作为输入,得到CDRs的结构
Steps:
- 用任意序列、位置和方向初始化CDR。扩散模型首先聚集了来自抗原和抗体框架的信息
- 迭代地更新cdr上每个氨基酸的氨基酸类型、位置和方向(侧链的方向)
- 我们基于预测的方向,使用侧链填充算法在原子级重建CDR结构
Detail
扩散过程
S : coordinates , X: amino acid types, O: orientations
- A diffusion probabilistic model defines two Markov chains of diffusion processes
- The forward diffusion process gradually adds noise to the data until the data distribution approximately reaches the prior distribution
- The generative diffusion process starts from the prior distribution and iteratively
transforms it to the desired distribution.
多项式分布 → 高斯分布
- 任意时刻 t t t,用 t 0 t_0 t0和 β \beta β表达
C C C:空间结构信息; R t R^t Rt :t时刻CDR的状态
all-one vector是一个元素全部为1的向量。例如,一个长度为n的all-one vector可以表示为[1, 1, 1, ..., 1]。在数学和计算机科学中,all-one vector经常用于矩阵和向量的运算和表示,例如在矩阵乘法中,一个矩阵乘以一个all-one vector可以得到该矩阵每一行的和。all-one vector也可以用于表示一组等权重的值,例如在计算平均值时,可以将每个值乘以一个all-one vector,再除以向量的长度,即可得到这组值的平均值。
- 可以用作去噪声
Diffusion For C a C_a Ca Coordinates :
- 坐标是一个正态分布
- 变化的学习率
- 神经网络用于预测高斯分布的噪声
- 目标函数是生成的分布和初始先验分布的MSE
SO(3) Denosing for Amino Acid Orientations:
- 各向同性的高斯分布,改变旋转角度
- 神经网络用于方向去噪和输出去噪的方向矩阵
- 目标函数是真实和预测的方向矩阵之间的差异内积
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-e8EAtnPL-1690184885993)(paper%20read%201%20035b71d4ad35406dbd00c2d05463cbd1/Untitled%2012.png)]
- For all Loss


![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-drIGTfju-1690184885987)(paper%20read%201%20035b71d4ad35406dbd00c2d05463cbd1/Untitled%204.png)]](http://img.e-com-net.com/image/info8/e0a82cd36af04c87a50926e61cd6a1f9.jpg)
![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-hGb3wfX6-1690184885989)(paper%20read%201%20035b71d4ad35406dbd00c2d05463cbd1/Untitled%205.png)]](http://img.e-com-net.com/image/info8/a3401ddb8ceb4b58a198ca94e1041fd1.jpg)


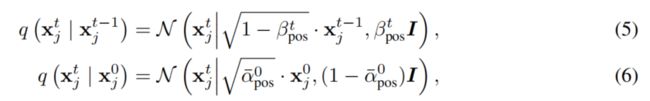
![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-FoE7p8iH-1690184885991)(paper%20read%201%20035b71d4ad35406dbd00c2d05463cbd1/Untitled%209.png)]](http://img.e-com-net.com/image/info8/36ea8515ec264ef4801789f077e996db.jpg)
![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-A0rsS9HK-1690184885992)(paper%20read%201%20035b71d4ad35406dbd00c2d05463cbd1/Untitled%2010.png)]](http://img.e-com-net.com/image/info8/9cb9e292df444be5b5fa1994f79ce59d.jpg)
![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-Qe87cPsQ-1690184885992)(paper%20read%201%20035b71d4ad35406dbd00c2d05463cbd1/Untitled%2011.png)]](http://img.e-com-net.com/image/info8/4fe98ab5e0614b38a22e6eba6fc5ed85.jpg)