PSP - 基于开源框架 OpenFold Multimer 蛋白质复合物的结构预测与BugFix
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/132410296
AlphaFold2-Multimer 是一个基于 AlphaFold2 的神经网络模型,可以预测多链蛋白复合物的结构。该模型在训练和推理时都可以处理多链输入,并且考虑了链之间的对称性和遗传信息。
- 对于 AlphaFold2 的损失函数、特征编码、裁剪策略和模型架构进行了多项修改,以适应多链蛋白复合物的特点。该模型还提供了一个基于预测 TM-score 的置信度评估方法。
- 在两个数据集上进行了评估,一个是 Benchmark 2,包含 17 个低同源性的异二聚体;另一个是 Recent-PDB-Multimers,包含 4,433 个最近的蛋白复合物。该模型使用 DockQ 分数来衡量预测结构与真实结构之间的接触质量。
- 在Benchmark 2上显著优于其他基于 AlphaFold2 或 ClusPro 的方法,在 Recent-PDB-Multimers 上也表现出较大的提升,尤其是在异构二聚体上。该模型还能够准确地预测自身的置信度,并且给出一些高质量的结构示例。
其中 OpenFold 是 AlphaFold2-Multimer 的开源实现,即:
- Paper: OpenFold: Retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization
- GitHub: https://github.com/aqlaboratory/openfold
将 OpenFold 的分支切换至 multimer 分支,即可使用 Multimer 功能,目前是 Debug 版本,基本推理功能已经支持,MSA 部分支持使用 AF2 的推理结果,模型支持 DeepMind 提供的 Multimer v3 模型,其余使用 OpenFold 的相关源码。评估当前 OpenFold Multimer框架的有效性。
其他参考文章:
- 开源可训练的蛋白质结构预测框架 OpenFold 的环境配置
- 基于 OpenFold 训练的 Finetuning 模型与推理逻辑评估
1. 模型效果
测试序列是 H1106_A122_B114.fasta,来源于 CASP15,即:
>A
MSRIITAPHIGIEKLSAISLEELSCGLPDRYALPPDGHPVEPHLERLYPTAQSKRSLWDFASPGYTFHGLHRAQDYRRELDTLQSLLTTSQSSELQAAAALLKCQQDDDRLLQIILNLLHKV
>B
MNITLTKRQQEFLLLNGWLQLQCGHAERACILLDALLTLNPEHLAGRRCRLVALLNNNQGERAEKEAQWLISHDPLQAGNWLCLSRAQQLNGDLDKARHAYQHYLELKDHNESP
OpenFold Multimer 的 MSA 文件夹格式,与 Monomer 类似,位于 alignments 文件夹中,不同的链放入同名文件夹中,即文件夹 A 和 B,具体文件如下:
bfd_uniref_hits.a3m
mgnify_hits.sto
pdb_hits.sto
uniprot_hits.sto
uniref90_hits.sto
其中 bfd_uniref_hits.a3m、mgnify_hits.sto、uniref90_hits.sto 是 MSA 的搜索结果,uniprot_hits.sto 用于 MSA Pairing,pdb_hits.sto 是模版搜索的结果。
测试命令,如下:
- 因为使用已有的 AlphaFold2 Multimer 搜索的 MSA,因此 MSA 相关配置并未启用;
- 模型使用 AF2 的
params_model_1_multimer_v3.npz,配置使用model_1_multimer_v3。
即:
python3 run_pretrained_openfold.py \
mydata/test-multimer \
af2-data-v230/pdb_mmcif/mmcif_files \
--uniref90_database_path af2-data-v230/uniref90/uniref90.fasta \
--mgnify_database_path af2-data-v230/mgnify/mgy_clusters_2022_05.fa \
--pdb70_database_path af2-data-v230/pdb70/pdb70 \
--uniclust30_database_path deepmsa2/uniclust30/uniclust30_2018_08 \
--uniref30_database_path af2-data-v230/uniref30/UniRef30_2021_03 \
--uniprot_database_path af2-data-v230/uniprot/uniprot.fasta \
--pdb_seqres_database_path af2-data-v230/pdb_seqres/pdb_seqres.txt \
--output_dir mydata/outputs-multimer/H1106_A122_B114/ \
--bfd_database_path af2-data-v230/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--model_device "cuda:0" \
--jackhmmer_binary_path /opt/openfold/hhsuite-speed/jackhmmer \
--hmmsearch_binary_path /opt/openfold/hhsuite-speed/hmmsearch \
--hhblits_binary_path /opt/conda/envs/openfold/bin/hhblits \
--hhsearch_binary_path /opt/conda/envs/openfold/bin/hhsearch \
--kalign_binary_path /opt/conda/envs/openfold/bin/kalign \
--config_preset "model_1_multimer_v3" \
--jax_param_path af2-data-v230/params/params_model_1_multimer_v3.npz \
--max_template_date 2022-04-01
运行日志如下,整体推理速度较快:
INFO:openfold/utils/script_utils.py:Successfully loaded JAX parameters at af2-data-v230/params/params_model_1_multimer_v3.npz...
INFO:run_pretrained_openfold.py:Using precomputed alignments for A at mydata/outputs-multimer/H1106_A122_B114/alignments...
INFO:run_pretrained_openfold.py:Using precomputed alignments for B at mydata/outputs-multimer/H1106_A122_B114/alignments...
INFO:openfold/utils/script_utils.py:Running inference for A-B...
INFO:openfold/utils/script_utils.py:Inference time: 44.876936707645655
INFO:run_pretrained_openfold.py:Output written to mydata/outputs-multimer/H1106_A122_B114/predictions/A-B_model_1_multimer_v3_unrelaxed.pdb...
INFO:run_pretrained_openfold.py:Running relaxation on mydata/outputs-multimer/H1106_A122_B114/predictions/A-B_model_1_multimer_v3_unrelaxed.pdb...
INFO:openfold/utils/script_utils.py:Relaxation time: 26.89977646060288
INFO:openfold/utils/script_utils.py:Relaxed output written to mydata/outputs-multimer/H1106_A122_B114/predictions/A-B_model_1_multimer_v3_relaxed.pdb...
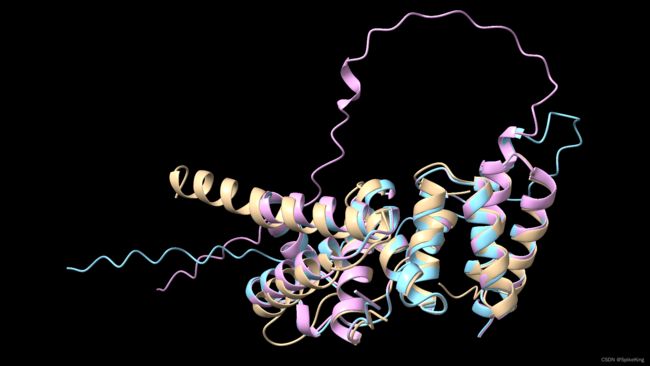
与 AlphaFold2 Multimer 的预测结果 unrelaxed_model_1_multimer_v3_pred_0.pdb,作为对比,效果在 H1106_A122_B114 中,略有提升,即:
[Info] {'TMScore': 0.8824, 'RMSD(local)': 1.92, 'Align.Len.': 173, 'DockQ': 0.613}
[Info] {'TMScore': 0.8803, 'RMSD(local)': 2.12, 'Align.Len.': 174, 'DockQ': 0.600}
其中,黄色是 Reference,蓝色是 AlphaFold2 Multimer 的预测结果,粉色是 OpenFold Multimer 的预测结果,如下:
2. Bugfix
Bug: 在MSA 序列 (sequence) 中,存在无法解析的 "." 关键字,导致 KeyError,即:
Traceback (most recent call last):
File "run_pretrained_openfold.py", line 477, in <module>
main(args)
File "run_pretrained_openfold.py", line 291, in main
feature_dict = generate_feature_dict(
File "run_pretrained_openfold.py", line 134, in generate_feature_dict
feature_dict = data_processor.process_fasta(
File "openfold/data/data_pipeline.py", line 1167, in process_fasta
chain_features = self._process_single_chain(
File "openfold/data/data_pipeline.py", line 1116, in _process_single_chain
chain_features = self._monomer_data_pipeline.process_fasta(
File "openfold/data/data_pipeline.py", line 860, in process_fasta
msa_features = self._process_msa_feats(alignment_dir, input_sequence, alignment_index)
File "openfold/data/data_pipeline.py", line 818, in _process_msa_feats
msa_features = make_msa_features(
File "openfold/data/data_pipeline.py", line 232, in make_msa_features
[residue_constants.HHBLITS_AA_TO_ID[res] for res in sequence]
File "openfold/data/data_pipeline.py", line 232, in <listcomp>
[residue_constants.HHBLITS_AA_TO_ID[res] for res in sequence]
KeyError: '.'
源码位于 openfold/data/data_pipeline.py 中,即:
def _process_msa_feats(
self,
alignment_dir: str,
input_sequence: Optional[str] = None,
alignment_index: Optional[str] = None
) -> Mapping[str, Any]:
msas = self._get_msas(
alignment_dir, input_sequence, alignment_index
)
msa_features = make_msa_features(
msas=msas
)
return msa_features
定义日志 logger,即:
import logging
logging.basicConfig()
logger = logging.getLogger(__file__)
logger.setLevel(level=logging.INFO)
定位 sequence,来源于 pdb_hits.sto 模版搜索结果,即:
INFO:openfold/data/data_pipeline.py:[CL] Error sequence: .MALLPDGQSI.EPHISR...LY...P....ERL.....ADRALLDFATPHR..GFHDLLRP.VD..FHQAMQ...G.LRSV.LAE.....GQSPELRAAA..ILLEQM.HADEQLMQMTLHLLHKV
原因:在 Multimer 中,Template 的搜索结果是 pdb_hits.sto,误解析成 MSA 文件,排除即可,同时,增加 pdb_hits.sto 的解析函数。
相关代码,各有 2 处,都需要修改,之前验证的是hmm_output,现修改成pdb_hits,即:
# ...
elif ext == ".sto" and "pdb_hits" not in filename:
msa = parsers.parse_stockholm(read_msa(start, size))
# ...
elif name == "pdb_hits.sto":
hits = parsers.parse_hmmsearch_sto(
read_template(start, size),
input_sequence,
)
all_hits[name] = hits
# ...