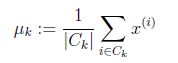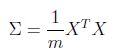Machine Learning week 8 programming exercise K-means Clustering and Principal Component Analysis
K-means Clustering
Implementing K-means
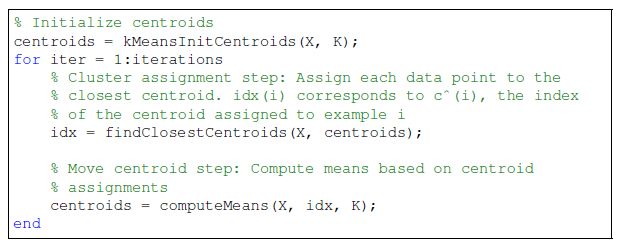
K-means 算法是一个循环算法,需要进行多次循环计算。并且有可能最后得到局部最优解,下面是算法的过程
第一步是cluster assignment,给每个training example找到最近的centroid点,然后确定分类index
function idx = findClosestCentroids(X, centroids)
%FINDCLOSESTCENTROIDS computes the centroid memberships for every example
% idx = FINDCLOSESTCENTROIDS (X, centroids) returns the closest centroids
% in idx for a dataset X where each row is a single example. idx = m x 1
% vector of centroid assignments (i.e. each entry in range [1..K])
%
% Set K
K = size(centroids, 1);
% You need to return the following variables correctly.
idx = zeros(size(X,1), 1);
% ====================== YOUR CODE HERE ======================
% Instructions: Go over every example, find its closest centroid, and store
% the index inside idx at the appropriate location.
% Concretely, idx(i) should contain the index of the centroid
% closest to example i. Hence, it should be a value in the
% range 1..K
%
% Note: You can use a for-loop over the examples to compute this.
%
X_k = idx; %选择这种算法是因为不想计算m,给了m的话,肯定就做2层循环了
for i = 1:K,
X_item = X - centroids(i, :);
X_item = X_item.*X_item;
X_item = sum(X_item, 2);%计算出每个点距离k中心点的距离平方
if i == 1,
X_k = X_item;
else
X_k = [X_k X_item];%最终每行排列的是K个数字,每个数字代表第i个点距离第k个点的距离平方
end;
end;
[val, ind] = min(X_k, [], 2); %返回每行的最小值和index
idx = ind;
% =============================================================
end
第二步是根据新的分类情况重新计算各个分类的centroid点,
function centroids = computeCentroids(X, idx, K)
%COMPUTECENTROIDS returs the new centroids by computing the means of the
%data points assigned to each centroid.
% centroids = COMPUTECENTROIDS(X, idx, K) returns the new centroids by
% computing the means of the data points assigned to each centroid. It is
% given a dataset X where each row is a single data point, a vector
% idx of centroid assignments (i.e. each entry in range [1..K]) for each
% example, and K, the number of centroids. You should return a matrix
% centroids, where each row of centroids is the mean of the data points
% assigned to it.
%
% Useful variables
[m n] = size(X);
% You need to return the following variables correctly.
centroids = zeros(K, n);
% ====================== YOUR CODE HERE ======================
% Instructions: Go over every centroid and compute mean of all points that
% belong to it. Concretely, the row vector centroids(i, :)
% should contain the mean of the data points assigned to
% centroid i.
%
% Note: You can use a for-loop over the centroids to compute this.
%
for i = 1:K,
k = find(idx==i);%注意这里不要写成一个等号,第一次就写错了
num = size(k, 1);
centroids(i,:) = sum(X(k,:),1)/num;
end;
% =============================================================
end
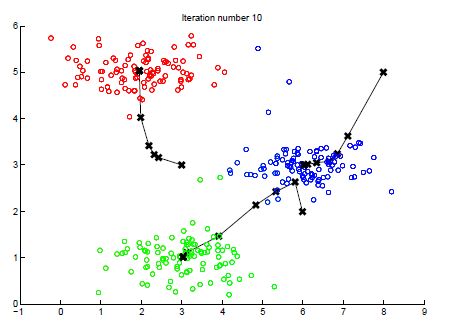
这里进行了10次循环,如果画出每次的变化过程的话
随机初始化,因为有可能会得到局部最优解,所以我们会多次随机初始化初始的centroid点,下面的代码进行了随机初始化,而且通过对training example的随机重新排列,并取其中前k个点,这样可以有效的避免重复元素的出现。
使用K-means算法压缩图像
这张图片的每个像素点都是有3个8bits的数字来分别代表Red,Green,Blue的颜色深度,也就是使用的RGB方法来进行的编码。这里我们使用K-means方法来压缩这个图片,通过k-means方法选出16个颜色,然后使用RGB方法表示这16个颜色,然后每个像素点只需要4bit就可以表示出来是16种元素的那个颜色。这样可以压缩到原来大小的1/6.
octave读图的方法
整个图片会对应一个三维的矩阵,其中前2维分别是点的横纵坐标,最后一维取值1-3,分别代表RGB中三种颜色的深度。扁平化处理之后,每个点会有3个features分别是rgb,然后在这3个feature上进行聚类,最终找到16个点。下图会是最后的结果,明显会有很多失真。
Principal Component Analysis
Implementing PCA
PCA最重要的有两个部分一个是计算covariance(sigma),另外是通过svd进行分解矩阵获得k个component,然后投影
当然进行PCA之前,我们有必要先对数据进行normalization,
sigma既covariance计算如下,代码十分简单
function [U, S] = pca(X) %PCA Run principal component analysis on the dataset X % [U, S, X] = pca(X) computes eigenvectors of the covariance matrix of X % Returns the eigenvectors U, the eigenvalues (on diagonal) in S % % Useful values [m, n] = size(X); % You need to return the following variables correctly. U = zeros(n); S = zeros(n); % ====================== YOUR CODE HERE ====================== % Instructions: You should first compute the covariance matrix. Then, you % should use the "svd" function to compute the eigenvectors % and eigenvalues of the covariance matrix. % % Note: When computing the covariance matrix, remember to divide by m (the % number of examples). % sigma = X'*X/m; [U, S, V] = svd(sigma); % ========================================================================= end
Projecting the data onto the principal components
去U的前K列,然后计算投影
function Z = projectData(X, U, K) %PROJECTDATA Computes the reduced data representation when projecting only %on to the top k eigenvectors % Z = projectData(X, U, K) computes the projection of % the normalized inputs X into the reduced dimensional space spanned by % the first K columns of U. It returns the projected examples in Z. % % You need to return the following variables correctly. Z = zeros(size(X, 1), K); % ====================== YOUR CODE HERE ====================== % Instructions: Compute the projection of the data using only the top K % eigenvectors in U (first K columns). % For the i-th example X(i,:), the projection on to the k-th % eigenvector is given as follows: % x = X(i, :)'; % projection_k = x' * U(:, k); % Z = X*U(:, 1:K); % ============================================================= end
下面是从k维恢复成n的方法
function X_rec = recoverData(Z, U, K) %RECOVERDATA Recovers an approximation of the original data when using the %projected data % X_rec = RECOVERDATA(Z, U, K) recovers an approximation the % original data that has been reduced to K dimensions. It returns the % approximate reconstruction in X_rec. % % You need to return the following variables correctly. X_rec = zeros(size(Z, 1), size(U, 1)); % ====================== YOUR CODE HERE ====================== % Instructions: Compute the approximation of the data by projecting back % onto the original space using the top K eigenvectors in U. % % For the i-th example Z(i,:), the (approximate) % recovered data for dimension j is given as follows: % v = Z(i, :)'; % recovered_j = v' * U(j, 1:K)'; % % Notice that U(j, 1:K) is a row vector. % X_rec = Z * U(:, 1:K)'; % ============================================================= end
PCA on Faces
老师给出了一组人脸的数据,分别是32x32的图片,那么我们会有的feature的数量将会是1024个
下面是PCA计算的前36个Component的图像显示
实验中使用了前100个component最为影射空间。然后把所有数据映射到新的100维空间中。这样可以大幅提升计算效率。
我们可以看一看在这个过程中我们损失了哪些信息,可以把数据从100维空间在转化为1024维的数据,当然这里面会有数据丢失,然后看下图,左边是原始数据,右边是恢复的数据。可见我们保留了基本的轮廓,但是细节部分都丢失了