数据挖掘实战(一)
这是做Kaggle的一个比赛的初步探索https://www.kaggle.com/c/reducing-commercial-aviation-fatalities
主要包括数据预处理、探索性分析、特征工程和建模四个步骤
1.数据预处理:
import warnings
import itertools
import numpy as np
import pandas as pd
import seaborn as sns
import lightgbm as lgb
import matplotlib.pyplot as plt
from tqdm import tqdm_notebook as tqdm
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, log_loss
warnings.simplefilter(action='ignore')
sns.set_style('whitegrid')读取数据和查看数据基本情况:
dtypes = {"crew": "int8",
"experiment": "category",
"time": "float32",
"seat": "int8",
"eeg_fp1": "float32",
"eeg_f7": "float32",
"eeg_f8": "float32",
"eeg_t4": "float32",
"eeg_t6": "float32",
"eeg_t5": "float32",
"eeg_t3": "float32",
"eeg_fp2": "float32",
"eeg_o1": "float32",
"eeg_p3": "float32",
"eeg_pz": "float32",
"eeg_f3": "float32",
"eeg_fz": "float32",
"eeg_f4": "float32",
"eeg_c4": "float32",
"eeg_p4": "float32",
"eeg_poz": "float32",
"eeg_c3": "float32",
"eeg_cz": "float32",
"eeg_o2": "float32",
"ecg": "float32",
"r": "float32",
"gsr": "float32",
"event": "category",
}
train_df = pd.read_csv("J:/CommercialAviationFatality/train.csv", dtype=dtypes)
test_df = pd.read_csv("J:/CommercialAviationFatality/test.csv", dtype=dtypes)
train_df.info()
train_df.head()
test_df.head()
探索性数据分析
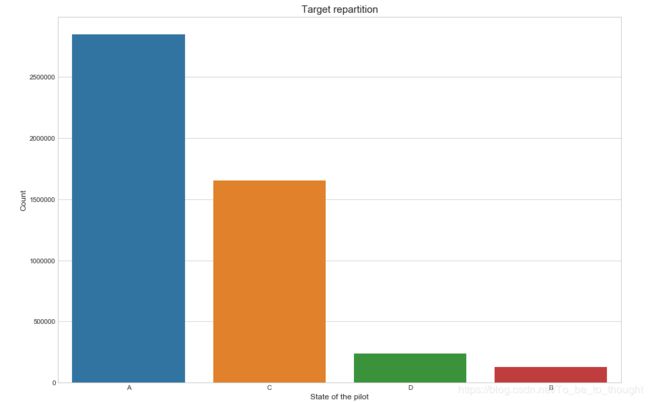
plt.figure(figsize=(15,10))
sns.countplot(train_df['event'])
plt.xlabel("State of the pilot", fontsize=12)
plt.ylabel("Count", fontsize=12)
plt.title("Target repartition", fontsize=15)
plt.show()plt.figure(figsize=(15,10))
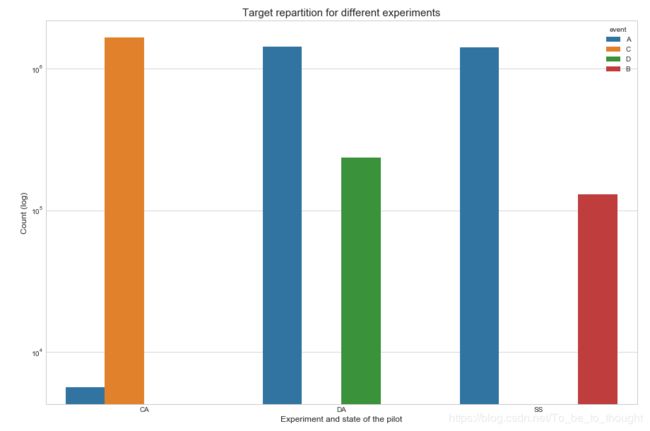
sns.countplot('experiment', hue='event', data=train_df)
plt.xlabel("Experiment and state of the pilot", fontsize=12)
plt.ylabel("Count (log)", fontsize=12)
plt.yscale('log')
plt.title("Target repartition for different experiments", fontsize=15)
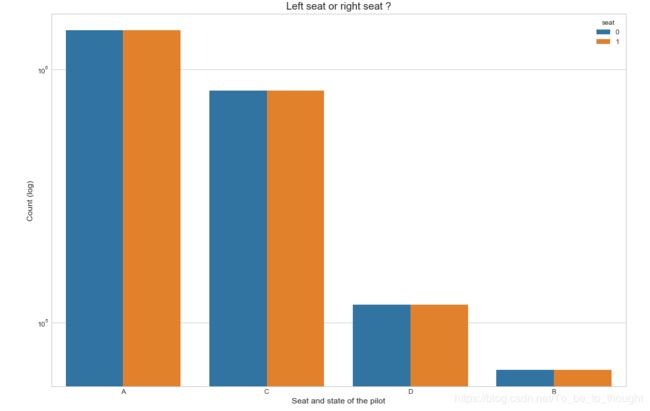
plt.show()plt.figure(figsize=(15,10))
sns.countplot('event', hue='seat', data=train_df)
plt.xlabel("Seat and state of the pilot", fontsize=12)
plt.ylabel("Count (log)", fontsize=12)
plt.yscale('log')
plt.title("Left seat or right seat ?", fontsize=15)
plt.show()
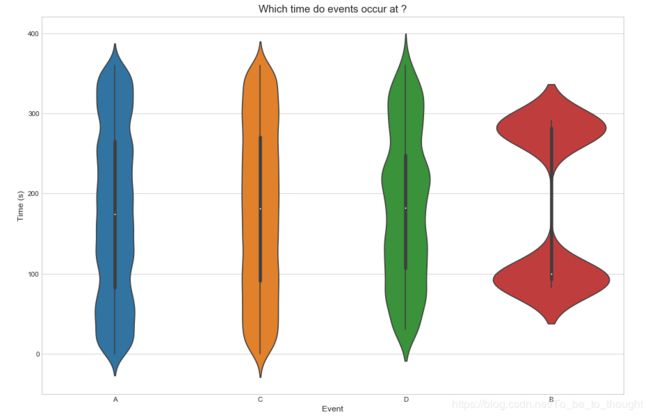
plt.figure(figsize=(15,10))
sns.violinplot(x='event', y='time', data=train_df.sample(50000))
plt.ylabel("Time (s)", fontsize=12)
plt.xlabel("Event", fontsize=12)
plt.title("Which time do events occur at ?", fontsize=15)
plt.show()plt.figure(figsize=(15,10))
sns.distplot(test_df['time'], label='Test set')
sns.distplot(train_df['time'], label='Train set')
plt.legend()
plt.xlabel("Time (s)", fontsize=12)
plt.title("Reparition of the time feature", fontsize=15)
plt.show()eeg_features = ["eeg_fp1", "eeg_f7", "eeg_f8", "eeg_t4", "eeg_t6", "eeg_t5", "eeg_t3", "eeg_fp2", "eeg_o1", "eeg_p3", "eeg_pz", "eeg_f3", "eeg_fz", "eeg_f4", "eeg_c4", "eeg_p4", "eeg_poz", "eeg_c3", "eeg_cz", "eeg_o2"]
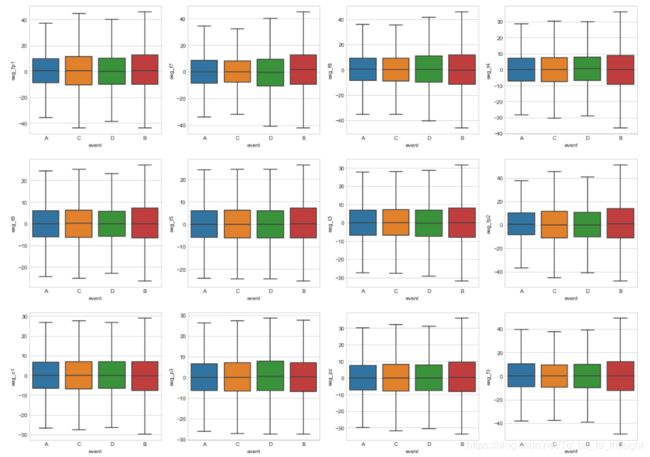
plt.figure(figsize=(20,25))
i = 0
for egg in eeg_features:
i += 1
plt.subplot(5, 4, i)
sns.boxplot(x='event', y=egg, data=train_df.sample(50000), showfliers=False)
plt.show()plt.figure(figsize=(20,25))
plt.title('Eeg features distributions')
i = 0
for eeg in eeg_features:
i += 1
plt.subplot(5, 4, i)
sns.distplot(test_df.sample(10000)[eeg], label='Test set', hist=False)
sns.distplot(train_df.sample(10000)[eeg], label='Train set', hist=False)
plt.xlim((-500, 500))
plt.legend()
plt.xlabel(eeg, fontsize=12)
plt.show()plt.figure(figsize=(15,10))
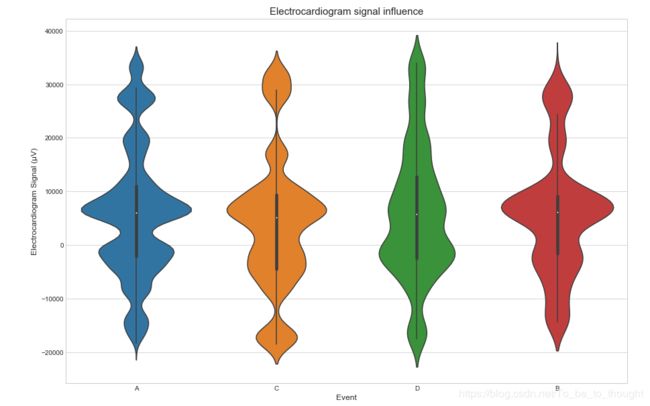
sns.violinplot(x='event', y='ecg', data=train_df.sample(50000))
plt.ylabel("Electrocardiogram Signal (µV)", fontsize=12)
plt.xlabel("Event", fontsize=12)
plt.title("Electrocardiogram signal influence", fontsize=15)
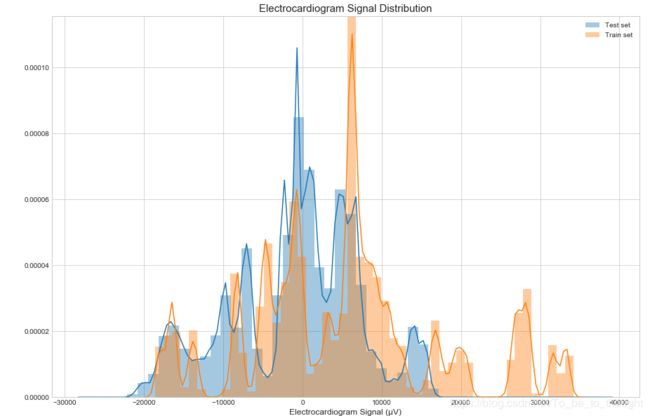
plt.show()plt.figure(figsize=(15,10))
sns.distplot(test_df['ecg'], label='Test set')
sns.distplot(train_df['ecg'], label='Train set')
plt.legend()
plt.xlabel("Electrocardiogram Signal (µV)", fontsize=12)
plt.title("Electrocardiogram Signal Distribution", fontsize=15)
plt.show()plt.figure(figsize=(15,10))
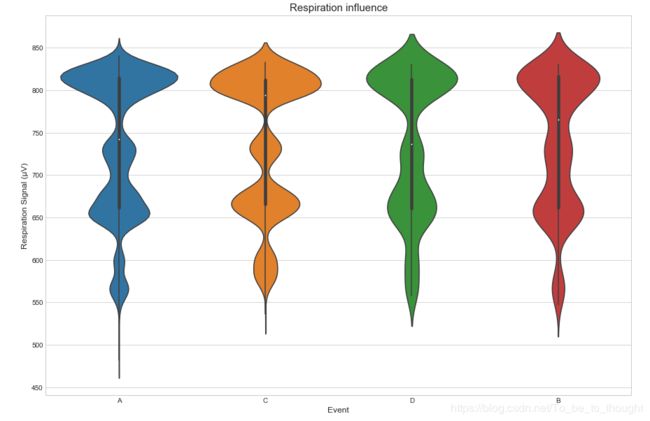
sns.violinplot(x='event', y='r', data=train_df.sample(50000))
plt.ylabel("Respiration Signal (µV)", fontsize=12)
plt.xlabel("Event", fontsize=12)
plt.title("Respiration influence", fontsize=15)
plt.show()plt.figure(figsize=(15,10))
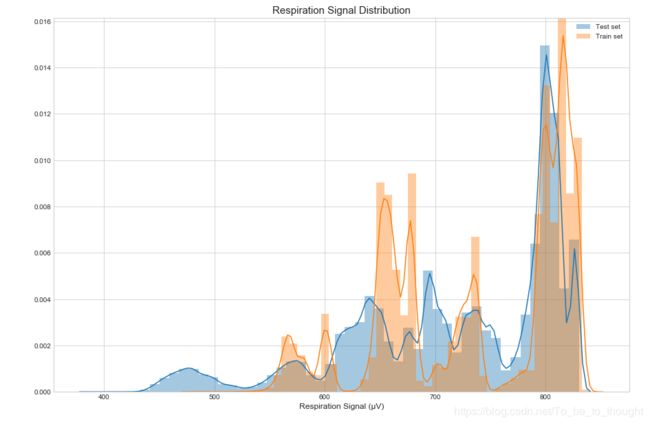
sns.distplot(test_df['r'], label='Test set')
sns.distplot(train_df['r'], label='Train set')
plt.legend()
plt.xlabel("Respiration Signal (µV)", fontsize=12)
plt.title("Respiration Signal Distribution", fontsize=15)
plt.show()plt.figure(figsize=(15,10))
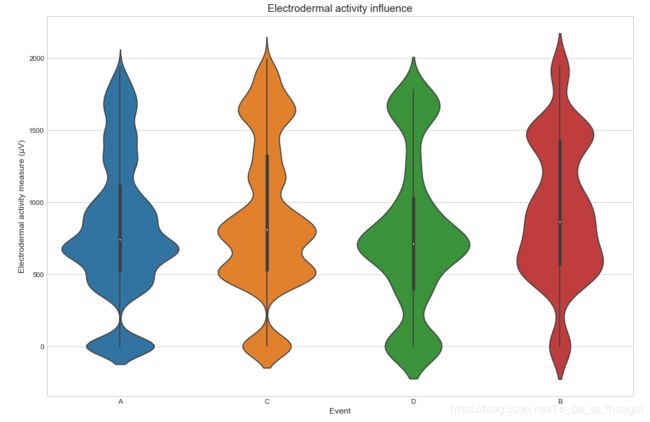
sns.violinplot(x='event', y='gsr', data=train_df.sample(50000))
plt.ylabel("Electrodermal activity measure (µV)", fontsize=12)
plt.xlabel("Event", fontsize=12)
plt.title("Electrodermal activity influence", fontsize=15)
plt.show()plt.figure(figsize=(15,10))
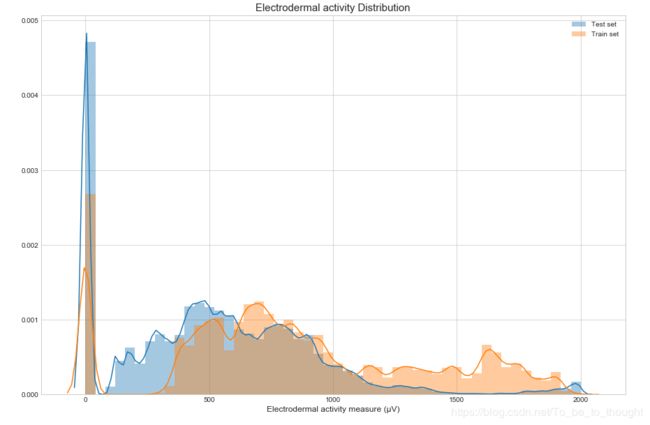
sns.distplot(test_df['gsr'], label='Test set')
sns.distplot(train_df['gsr'], label='Train set')
plt.legend()
plt.xlabel("Electrodermal activity measure (µV)", fontsize=12)
plt.title("Electrodermal activity Distribution", fontsize=15)
plt.show()features_n = ["eeg_fp1", "eeg_f7", "eeg_f8", "eeg_t4", "eeg_t6", "eeg_t5", "eeg_t3", "eeg_fp2", "eeg_o1", "eeg_p3", "eeg_pz", "eeg_f3", "eeg_fz", "eeg_f4", "eeg_c4", "eeg_p4", "eeg_poz", "eeg_c3", "eeg_cz", "eeg_o2", "ecg", "r", "gsr"]
train_df['pilot'] = 100 * train_df['seat'] + train_df['crew']
test_df['pilot'] = 100 * test_df['seat'] + test_df['crew']
print("Number of pilots : ", len(train_df['pilot'].unique()))
def normalize_by_pilots(df):
pilots = df["pilot"].unique()
for pilot in tqdm(pilots):
ids = df[df["pilot"] == pilot].index
scaler = MinMaxScaler()
df.loc[ids, features_n] = scaler.fit_transform(df.loc[ids, features_n])
return df
train_df = normalize_by_pilots(train_df)
test_df = normalize_by_pilots(test_df)
train_df, val_df = train_test_split(train_df, test_size=0.2, random_state=420)
print(f"Training on {train_df.shape[0]} samples.")
features = ["crew", "seat"] + features_n
def run_lgb(df_train, df_test):
# Classes as integers
dic = {'A': 0, 'B': 1, 'C': 2, 'D': 3}
try:
df_train["event"] = df_train["event"].apply(lambda x: dic[x])
df_test["event"] = df_test["event"].apply(lambda x: dic[x])
except:
pass
params = {"objective" : "multiclass",
"num_class": 4,
"metric" : "multi_error",
"num_leaves" : 30,
"min_child_weight" : 50,
"learning_rate" : 0.1,
"bagging_fraction" : 0.7,
"feature_fraction" : 0.7,
"bagging_seed" : 420,
"verbosity" : -1
}
lg_train = lgb.Dataset(df_train[features], label=(df_train["event"]))
lg_test = lgb.Dataset(df_test[features], label=(df_test["event"]))
model = lgb.train(params, lg_train, 1000, valid_sets=[lg_test], early_stopping_rounds=50, verbose_eval=100)
return model
model = run_lgb(train_df, val_df)
fig, ax = plt.subplots(figsize=(12,10))
lgb.plot_importance(model, height=0.8, ax=ax)
ax.grid(False)
plt.ylabel('Feature', size=12)
plt.xlabel('Importance', size=12)
plt.title("Importance of the Features of our LightGBM Model", fontsize=15)
plt.show()
pred_val = model.predict(val_df[features], num_iteration=model.best_iteration)
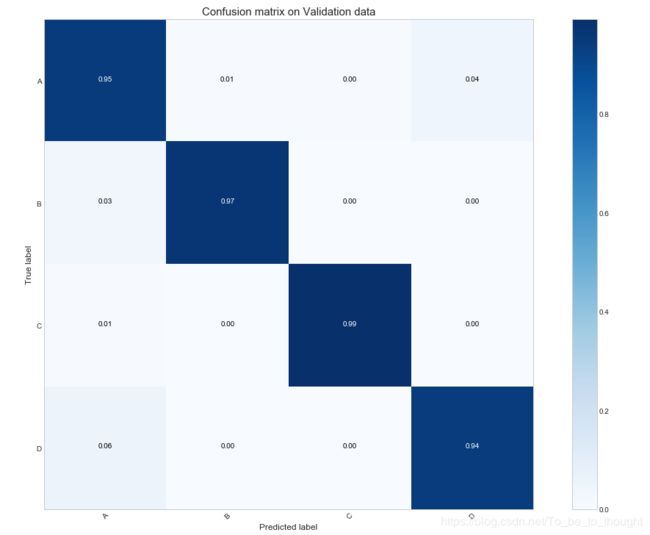
print("Log loss on validation data :", round(log_loss(np.array(val_df["event"].values), pred_val), 3))def plot_confusion_matrix(cm, classes, title='Confusion matrix', normalize=False, cmap=plt.cm.Blues):
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
fmt = '.2f' if normalize else 'd'
fig, ax = plt.subplots(figsize=(15, 10))
ax.imshow(cm, interpolation='nearest', cmap=cmap)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title, size=15)
plt.colorbar()
plt.grid(False)
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
thresh = (cm.max()+cm.min()) / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label', size=12)
plt.xlabel('Predicted label', size=12)
conf_mat_val = confusion_matrix(np.argmax(pred_val, axis=1), val_df["event"].values)
plot_confusion_matrix(conf_mat_val, ["A", "B", "C", "D"], title='Confusion matrix on Validation data', normalize=True)