GSEA支持的数据格式
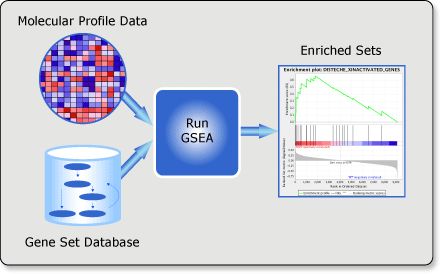
Gene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes).
官网给出的数据格式:
Data formats
- 1 Expression Data Formats
- 1.1 GCT: Gene Cluster Text file format (*.gct)
- 1.2 RES: ExpRESsion (with P and A calls) file format (*.res)
- 1.3 PCL: Stanford cDNA file format (*.pcl)
- 1.4 TXT: Text file format for expression dataset (*.txt)
- 2 Phenotype Data Formats
- 2.1 CLS: Categorical (e.g tumor vs normal) class file format (*.cls)
- 2.2 CLS: Continuous (e.g time-series or gene profile) file format (*.cls)
- 3 Gene Set Database Formats
- 3.1 GMX: Gene MatriX file format (*.gmx)
- 3.2 GMT: Gene Matrix Transposed file format (*.gmt)
- 3.3 GRP: Gene set file format (*.grp)
- 3.4 XML: Molecular signature database file format (msigdb_*.xml)
- 4 Microarray Chip Annotation Formats
- 4.1 CHIP: Chip file format (*.chip)
- 5 Ranked Gene Lists
- 5.1 RNK: Ranked list file format (*.rnk)
1 Expression Data Formats
1.1 GCT: Gene Cluster Text file format (*.gct)
tab键分割的文档
第一行:版本信息
#1.2
第二行:两个数字描述数据的大小,第一个数字是行数(不包括前三行),第二个数字是列数(不包括前两列:名字和描述)
Line format: (# of data rows) (tab) (# of data columns)
Example:
7129 58
第三行:每列的列名
Line format: Name(tab)Description(tab)(sample 1 name)(tab)(sample 2 name) (tab) ... (sample N name)
Example: Name Description DLBC1_1 DLBC2_1 ... DLBC58_0
剩下的数据:基因相关的信息,每行是一个基因,每列是一个样品;行数与列数与应该与第二行设定的数字一样;每行都有一个名字,描述信息,每个样品对应一个数值;列名与描述信息可以包含空格,但是不能为空,描述信息如果为空,就添加NA or NULL;
Line format: (gene name) (tab) (gene description) (tab) (col 1 data) (tab) (col 2 data) (tab) ... (col N data)
Example: AFFX-BioB-5_at AFFX-BioB-5_at (endogenous control) -104 -152 -158 ... -44
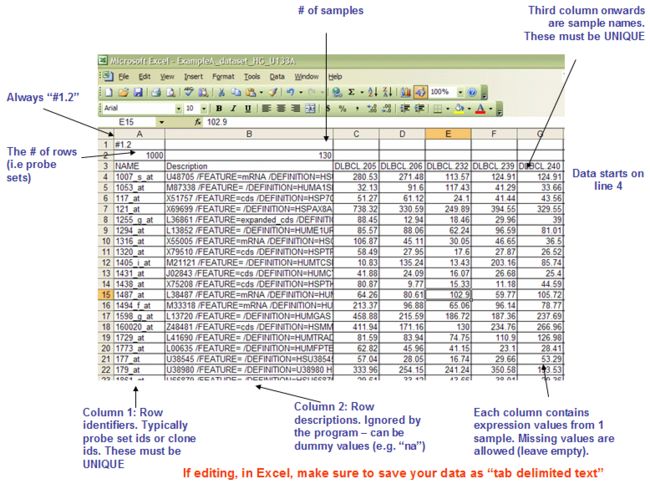
1.4 TXT: Text file format for expression dataset (*.txt)
tab键分割的文档
第一行:
Line format: Name(tab)Description(tab)(sample 1 name)(tab)(sample 2 name) (tab) ... (sample N name)
Example: Name Description DLBC1_1 DLBC2_1 ... DLBC58_0
.txt格式文档就是.gct格式文档直接取出前三行;
2 Phenotype Data Formats
表型文档定义了表达文档中样品的表型标签,使用空格或tab隔开;表型文档分为类别型与连续性;
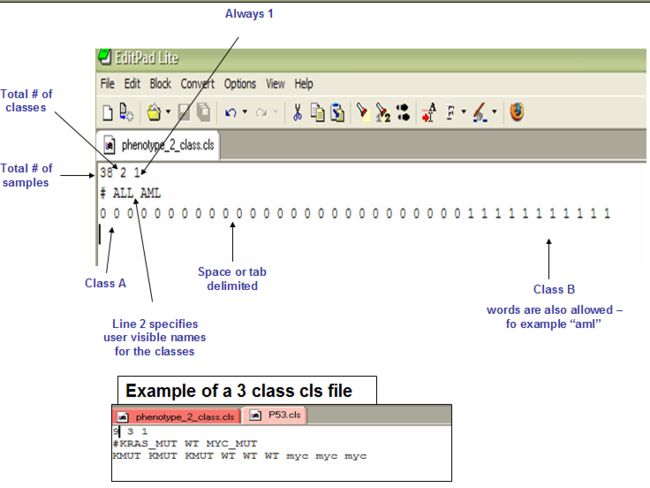
2.1 CLS: Categorical (e.g tumor vs normal) class file format (.cls)
第一行:样品个数,类别
Line format: (number of samples) (space) (number of classes) (space) 1
Example: 58 2 1
第二行:类别名,#开始
Line format: # (space) (class 0 name) (space) (class 1 name)
Example: # cured fatal/ref
第三行:每个样品的一个类标签,可以定义为类名,数字或字符串;类标签的顺序必须与第二行的类名顺序一致(虽然每个类标签都重复);类标签必须与表达数据中的样品名一一对应;
Line format: (sample 1 class) (space) (sample 2 class) (space) ... (sample N class)
Example: 0 0 0 ... 1 1
CLS: Continuous (e.g time-series or gene profile) file format (.cls)
连续性表型文件包含一个或多个类别;
#numeric
#AFFX-BioB-5_st
206.0 31.0 252.0 -20.0 -169.0 -66.0 230.0 -23.0 67.0 173.0 -55.0 -20.0 469.0 -201.0 -117.0 -162.0 -5.0 -86.0 350.0 74.0 -215.0 193.0 506.0 183.0 350.0 113.0 -17.0 29.0 247.0 -131.0 358.0 561.0 24.0 524.0 167.0 -56.0 176.0 320.0
#AFFX-BioDn-5
75.0 142.0 32.0 109.0 -38.0 -80.0 62.0 39.0 196.0 -42.0 199.0 49.0 171.0 327.0 115.0 -71.0 85.0 80.0 270.0 182.0 208.0 -94.0 292.0 233.0 34.0 0.0 59.0 233.0 48.0 466.0 -7.0 -96.0 297.0 38.0 208.0 -15.0 30.0 357.0
第一行:"#numeric"指明这是一个连续性标签
剩下的数据就是连续性标签表型:
每两行定义一个分类:
第一行:类别名,如 #AFFX-BIOB-5_st
第二行:一行对应表达数据中每个样品的值
For a continuous phenotype label, the values for the samples define the phenotype profile. The relative change in the values defines the relative distance between points in the phenotype profile. In the example shown above, the sample values for the two phenotype labels are gene expression values. The phenotype profile is the expression profile for a gene and is used to find gene sets correlated with that gene. For a time series experiment, you would choose sample values that define the desired expression profile. The example shown below assumes that you have five samples taken at 30 minute intervals. The first phenotype label defines a phenotype profile that shows steadily increasing gene expression; the second defines a profile that shows an initial peak and then gradual decrease:
#numeric
- IncreasingProfle
30 60 90 120 150
- PeakProfle
5 20 15 10 5
3 Gene Set Database Formats
通常有两种格式:
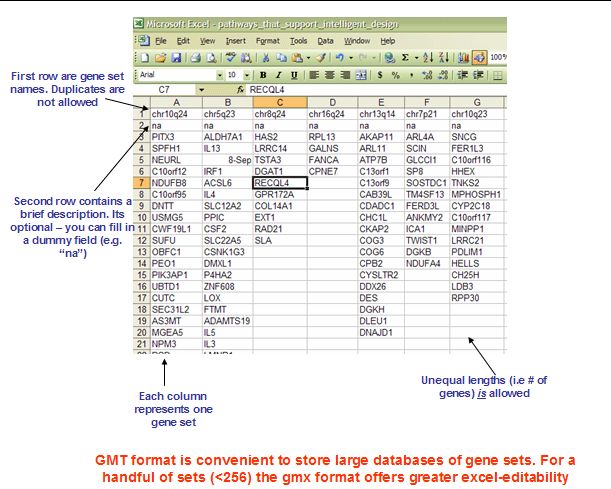
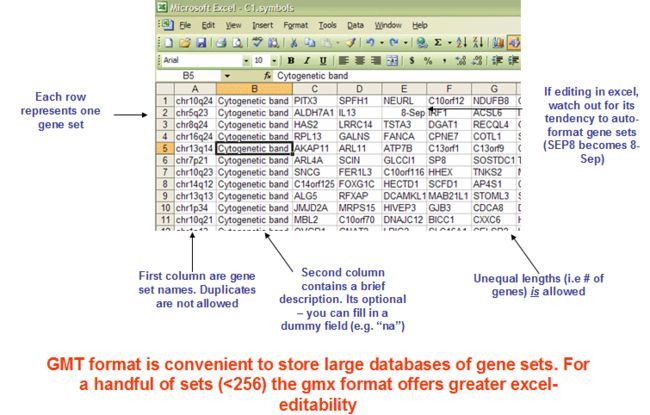
GMX中每列是一个基因集,包含名字,描述,基因集中的基因;
GMT中每行是一个基因集,包含名字,描述,基因集中的基因;
3.1 GMX: Gene MatriX file format (*.gmx)
3.2 GMT: Gene Matrix Transposed file format (*.gmt)
4 Microarray Chip Annotation Formats
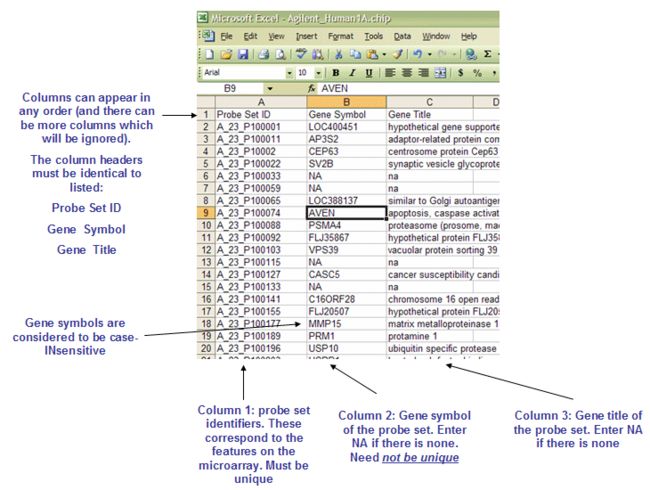
4.1 CHIP: Chip file format (*.chip)
文件中包含了对芯片的注释;
第一行是每一列的列名:
文件中必须包含的三列:Probe Set ID Gene Symbol Gene Title
The GENE_SYMBOL.chip file contains one additional column, Aliases, which is not shown here. When a gene is identified by more than one HUGO gene symbol, the Gene Symbol column contains the gene symbol that appears in the GSEA reports and the Alias column identifies other gene symbols used to reference the same gene. If a gene set or chip annotation file contains a gene in the Alias column, GSEA automatically converts it to the gene in the Gene Symbol column.
The rest of the file contains data for each probe set ID used in the microarray.
Line format: (probe set id) (tab) (gene symbol) (tab) (gene title)
5 Ranked Gene Lists
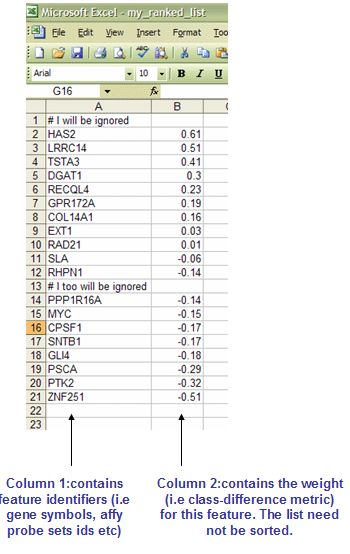
5.1 RNK: Ranked list file format (*.rnk)
RNK这种格式文件中存放的是可排序的基因列表。下图就是一个例子展示,第一列是基因名,第二列是用于基因列表排序统计学数值。