局部加权回归(LOWESS)
文章目录
- 核函数
- 叶帕涅奇尼科夫(epanechnikov)核函数
- 立方核
- 应用核函数
- 局部加权回归
- 增强鲁棒性
- 双平方函数
- 鲁棒局部加权线性回归
局部加权线性回归:local weighted regression
核函数
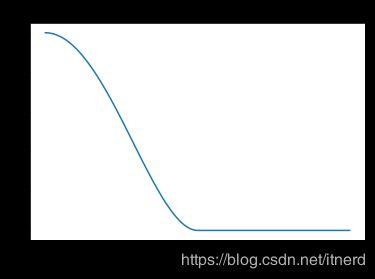
叶帕涅奇尼科夫(epanechnikov)核函数
又称二次核
K l ( x , x 0 ) = D ( ∣ ∣ x − x 0 ∣ ∣ l ) K_l(x,x_0) = D(\frac{||x-x_0||}{l}) Kl(x,x0)=D(l∣∣x−x0∣∣)
D ( t ) = { 0.75 ( 1 − t 2 ) , ∣ t ∣ < 1 , 0 , o t h e r w i s e . D(t) = \left\{ \begin{array}{ll} 0.75(1-t^2), & |t|<1,\\ 0, & otherwise. \end{array} \right. D(t)={0.75(1−t2),0,∣t∣<1,otherwise.
# Kernel functions:
def epanechnikov(xx, **kwargs):
"""
The Epanechnikov kernel estimated for xx values at indices idx (zero
elsewhere)
Parameters
----------
xx: float array
Values of the function on which the kernel is computed. Typically,
these are Euclidean distances from some point x0 (see do_kernel)
"""
l = kwargs.get('l', 1.0)
ans = np.zeros(xx.shape)
xx_norm = xx / l
idx = np.where(xx_norm <= 1)
ans[idx] = 0.75 * (1 - xx_norm[idx] ** 2)
return ans
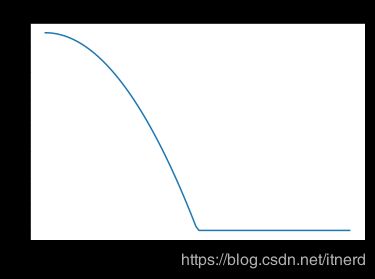
立方核
K l ( x , x 0 ) = D ( ∣ ∣ x − x 0 ∣ ∣ l ) K_l(x,x_0) = D(\frac{||x-x_0||}{l}) Kl(x,x0)=D(l∣∣x−x0∣∣)
D ( t ) = { ( 1 − ∣ t ∣ 3 ) 3 , ∣ t ∣ < 1 , 0 , o t h e r w i s e . D(t) = \left\{ \begin{array}{ll} (1-|t|^3)^3, & |t|<1,\\ 0, & otherwise. \end{array} \right. D(t)={(1−∣t∣3)3,0,∣t∣<1,otherwise.
def tri_cube(xx, **kwargs):
"""
The tri-cube kernel estimated for xx values at indices idx (zero
elsewhere)
Parameters
----------
xx: float array
Values of the function on which the kernel is computed. Typically,
these are Euclidean distances from some point x0 (see do_kernel)
idx: tuple
An indexing tuple pointing to the coordinates in xx for which the
kernel value is estimated. Default: None (all points are used!)
"""
ans = np.zeros(xx.shape)
idx = np.where(xx <= 1)
ans[idx] = (1 - np.abs(xx[idx]) ** 3) ** 3
return ans
应用核函数
d o _ k e r n e l = k e r n e l ( ∣ ∣ x − x 0 ∣ ∣ 2 ; l ) do\_kernel = kernel(||x-x_0||_2;l) do_kernel=kernel(∣∣x−x0∣∣2;l)
def do_kernel(x0, x, l=1.0, kernel=epanechnikov):
"""
Calculate a kernel function on x in the neighborhood of x0
Parameters
----------
x: float array
All values of x
x0: float
The value of x around which we evaluate the kernel
l: float or float array (with shape = x.shape)
Width parameter (metric window size)
kernel: callable
A kernel function. Any function with signature: `func(xx)`
"""
# xx is the norm of x-x0. Note that we broadcast on the second axis for the
# nd case and then sum on the first to get the norm in each value of x:
xx = np.sum(np.sqrt(np.power(x - x0[:, np.newaxis], 2)), 0)
return kernel(xx, l=l)
局部加权回归
拟合函数
f ( x ) = ∑ i = 0 d α i x i f(x) = \sum_{i=0}^d \alpha_i x^i f(x)=i=0∑dαixi
优化目标
min α ∑ i = 1 N K ( x , x i ) ( y i − ∑ j = 0 d α j x j ) 2 \min_\alpha \quad \sum_{i=1}^N K(x,x_i) \left(y_i - \sum_{j=0}^d \alpha_j x^j \right)^2 αmini=1∑NK(x,xi)(yi−j=0∑dαjxj)2
写成矩阵形式
min A ( Y − X A ) ⊤ K ( Y − X A ) \min_A \quad (Y-XA)^\top K(Y-XA) Amin(Y−XA)⊤K(Y−XA)
解得:
A = ( X ⊤ K X ) − 1 X ⊤ K Y A = (X^\top KX)^{-1}X^\top KY A=(X⊤KX)−1X⊤KY
增强鲁棒性
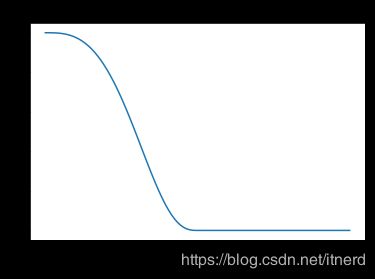
双平方函数
用双平方函数来降低异常点的影响,见下一节。
双平方函数为
B ( t ) = { ( 1 − t 2 ) 2 , ∣ t ∣ < 1 , 0 , o t h e r w i s e . B(t) = \left\{ \begin{array}{ll} (1-t^2)^2, & |t|<1,\\ 0, & otherwise. \end{array} \right. B(t)={(1−t2)2,0,∣t∣<1,otherwise.
def bi_square(xx, **kwargs):
"""
The bi-square weight function calculated over values of xx
Parameters
----------
xx: float array
"""
ans = np.zeros(xx.shape)
idx = np.where(xx < 1)
ans[idx] = (1 - xx[idx] ** 2) ** 2
return ans
鲁棒局部加权线性回归
先做一次局部加权回归,计算出拟合的残差:
e i = y i − f ( x i ) e_i = y_i - f(x_i) ei=yi−f(xi)
利用双平方函数计算误差权重:
δ i = B ( e i / 6 s ) \delta_i = B(e_i/6s) δi=B(ei/6s)
其中 s s s 为误差绝对值 ∣ e i ∣ |e_i| ∣ei∣ 的中位数,上式的含义是将误差大于6倍中位数的点视为离群点,权重赋为 0.
将 δ i \delta_i δi 乘以核函数得到的距离权重 K ( x , x i ) K(x,x_i) K(x,xi),再做一次加权回归。
def lowess(x, y, x0, deg=1, kernel=epanechnikov, l=1, robust=False,):
"""
Locally smoothed regression with the LOWESS algorithm.
Parameters
----------
x: float n-d array
Values of x for which f(x) is known (e.g. measured). The shape of this
is (n, j), where n is the number the dimensions and j is the
number of distinct coordinates sampled.
y: float array
The known values of f(x) at these points. This has shape (j,)
x0: float or float array.
Values of x for which we estimate the value of f(x). This is either a
single scalar value (only possible for the 1d case, in which case f(x0)
is estimated for just that one value of x), or an array of shape (n, k).
deg: int
The degree of smoothing functions. 0 is locally constant, 1 is locally
linear, etc. Default: 1.
kernel: callable
A kernel function. {'epanechnikov', 'tri_cube', 'bi_square'}
l: float or float array with shape = x.shape
The metric window size for the kernel
robust: bool
Whether to apply the robustification procedure from [Cleveland79], page
831
Returns
-------
The function estimated at x0.
"""
if robust:
# We use the procedure described in Cleveland1979
# Start by calling this function with robust set to false and the x0
# input being equal to the x input:
y_est = lowess(x, y, x, kernel=epanechnikov, l=l, robust=False)
resid = y_est - y
median_resid = np.nanmedian(np.abs(resid))
# Calculate the bi-cube function on the residuals for robustness
# weights:
robustness_weights = bi_square(resid / (6 * median_resid))
# For the case where x0 is provided as a scalar:
if not np.iterable(x0):
x0 = np.asarray([x0])
ans = np.zeros(x0.shape[-1])
# We only need one design matrix for fitting:
B = [np.ones(x.shape[-1])]
for d in range(1, deg+1):
B.append(x ** deg)
B = np.vstack(B).T
for idx, this_x0 in enumerate(x0.T):
# This is necessary in the 1d case (?):
if not np.iterable(this_x0):
this_x0 = np.asarray([this_x0])
# Different weighting kernel for each x0:
W = np.diag(do_kernel(this_x0, x, l=l, kernel=kernel))
# XXX It should be possible to calculate W outside the loop, if x0 and
# x are both sampled in some regular fashion (that is, if W is the same
# matrix in each iteration). That should save time.
if robust:
# We apply the robustness weights to the weighted least-squares
# procedure:
robustness_weights[np.isnan(robustness_weights)] = 0
W = np.dot(W, np.diag(robustness_weights))
BtWB = np.dot(np.dot(B.T, W), B)
BtW = np.dot(B.T, W)
# Get the params:
beta = np.dot(np.dot(la.pinv(BtWB), BtW), y.T)
# We create a design matrix for this coordinat for back-predicting:
B0 = [1]
for d in range(1, deg+1):
B0 = np.hstack([B0, this_x0 ** deg])
B0 = np.vstack(B0).T
# Estimate the answer based on the parameters:
ans[idx] += np.dot(B0, beta)
# If we are trying to sample far away from where the function is
# defined, we will be trying to invert a singular matrix. In that case,
# the regression should not work for you and you should get a nan:
#except la.LinAlgError :
# ans[idx] += np.nan
return ans.T